Initial commit
This commit is contained in:
parent
39c5f79e55
commit
d843ca2c7f
|
|
@ -0,0 +1,31 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Tue Jun 11 17:10:43 2019
|
||||||
|
|
||||||
|
@author: Administrator
|
||||||
|
"""
|
||||||
|
|
||||||
|
|
||||||
|
import cv2
|
||||||
|
import numpy as np
|
||||||
|
|
||||||
|
|
||||||
|
img = cv2.imread('a.png',0)
|
||||||
|
img1 = img.astype('float')
|
||||||
|
|
||||||
|
def dct(m):
|
||||||
|
m = np.float32(m)/255.0
|
||||||
|
return cv2.dct(m)*255
|
||||||
|
#print(dct(img1).shape)
|
||||||
|
new_dct=dct(img1)
|
||||||
|
after_dct=[]
|
||||||
|
for i in range(len(new_dct)):
|
||||||
|
for j in range(len(new_dct[0])):
|
||||||
|
after_dct.append(int(new_dct[i][j]))
|
||||||
|
#print(new_dct)
|
||||||
|
#new_dct=new_dct.reshape(-1,1)
|
||||||
|
#print(len(after_dct))
|
||||||
|
#print(after_dct[:600])
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
@ -0,0 +1,60 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Mon Jun 10 17:30:40 2019
|
||||||
|
|
||||||
|
@author: Administrator
|
||||||
|
"""
|
||||||
|
|
||||||
|
from Jsteg import Jsteg
|
||||||
|
|
||||||
|
|
||||||
|
class F3(Jsteg):
|
||||||
|
def __init__(self):
|
||||||
|
Jsteg.__init__(self)
|
||||||
|
|
||||||
|
def set_sequence_after_dct(self,sequence_after_dct):
|
||||||
|
self.sequence_after_dct=sequence_after_dct
|
||||||
|
sum_len=len(self.sequence_after_dct)
|
||||||
|
zero_len=len([i for i in self.sequence_after_dct if i==0])
|
||||||
|
one_len=len([i for i in self.sequence_after_dct if i in (-1,1)])
|
||||||
|
self.available_info_len=sum_len-zero_len-one_len # 不是特别可靠
|
||||||
|
print ("Load>> 大约可嵌入",sum_len-zero_len-int(one_len/2),'bits')
|
||||||
|
print ("Load>> 最少可嵌入",self.available_info_len,'bits\n')
|
||||||
|
|
||||||
|
def _write(self,index,data):
|
||||||
|
origin=self.sequence_after_dct[index]
|
||||||
|
if origin == 0:
|
||||||
|
return False
|
||||||
|
elif origin in (-1,1) and data==0:
|
||||||
|
self.sequence_after_dct[index]=0
|
||||||
|
return False
|
||||||
|
|
||||||
|
lower_bit=origin%2
|
||||||
|
|
||||||
|
if lower_bit==data:
|
||||||
|
pass
|
||||||
|
elif origin>0:
|
||||||
|
self.sequence_after_dct[index]=origin-1
|
||||||
|
elif origin<0:
|
||||||
|
self.sequence_after_dct[index]=origin+1
|
||||||
|
return True
|
||||||
|
|
||||||
|
def _read(self,index):
|
||||||
|
if self.sequence_after_dct[index] != 0:
|
||||||
|
return self.sequence_after_dct[index]%2
|
||||||
|
else:
|
||||||
|
return None
|
||||||
|
|
||||||
|
|
||||||
|
if __name__=="__main__":
|
||||||
|
f3=F3()
|
||||||
|
# 写
|
||||||
|
sequence_after_dct=[-1,0,1]*100+[i for i in range(-7,500)]
|
||||||
|
f3.set_sequence_after_dct(sequence_after_dct)
|
||||||
|
info1=[0,1,0,1,1,0,1,0]
|
||||||
|
f3.write(info1)
|
||||||
|
# 读
|
||||||
|
sequence_after_dct2=f3.get_sequence_after_dct()
|
||||||
|
f3.set_sequence_after_dct(sequence_after_dct2)
|
||||||
|
info2=f3.read()
|
||||||
|
print (info2)
|
||||||
|
|
@ -0,0 +1,67 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Tue Jun 11 16:10:51 2019
|
||||||
|
|
||||||
|
@author: Administrator
|
||||||
|
"""
|
||||||
|
|
||||||
|
from Jsteg import Jsteg
|
||||||
|
|
||||||
|
|
||||||
|
class F4(Jsteg):
|
||||||
|
def __init__(self):
|
||||||
|
Jsteg.__init__(self)
|
||||||
|
|
||||||
|
def set_sequence_after_dct(self,sequence_after_dct):
|
||||||
|
self.sequence_after_dct=sequence_after_dct
|
||||||
|
sum_len=len(self.sequence_after_dct)
|
||||||
|
zero_len=len([i for i in self.sequence_after_dct if i==0])
|
||||||
|
one_len=len([i for i in self.sequence_after_dct if i in (-1,1)])
|
||||||
|
self.available_info_len=sum_len-zero_len-one_len # 不是特别可靠
|
||||||
|
print ("Load>> 大约可嵌入",sum_len-zero_len-int(one_len/2),'bits')
|
||||||
|
print ("Load>> 最少可嵌入",self.available_info_len,'bits\n')
|
||||||
|
|
||||||
|
def _write(self,index,data):
|
||||||
|
origin=self.sequence_after_dct[index]
|
||||||
|
if origin == 0:
|
||||||
|
return False
|
||||||
|
elif origin == 1 and data==0:
|
||||||
|
self.sequence_after_dct[index]=0
|
||||||
|
return False
|
||||||
|
|
||||||
|
elif origin == -1 and data==1:
|
||||||
|
self.sequence_after_dct[index]=0
|
||||||
|
return False
|
||||||
|
|
||||||
|
lower_bit=origin%2
|
||||||
|
|
||||||
|
if origin >0:
|
||||||
|
if lower_bit!=data:
|
||||||
|
self.sequence_after_dct[index]=origin-1
|
||||||
|
else:
|
||||||
|
if lower_bit==data:
|
||||||
|
self.sequence_after_dct[index]=origin+1
|
||||||
|
return True
|
||||||
|
|
||||||
|
|
||||||
|
def _read(self,index):
|
||||||
|
if self.sequence_after_dct[index] >0:
|
||||||
|
return self.sequence_after_dct[index]%2
|
||||||
|
elif self.sequence_after_dct[index]<0:
|
||||||
|
return (self.sequence_after_dct[index]+1)%2
|
||||||
|
else:
|
||||||
|
return None
|
||||||
|
|
||||||
|
|
||||||
|
if __name__=="__main__":
|
||||||
|
f4=F4()
|
||||||
|
# 写
|
||||||
|
sequence_after_dct=[-1,0,1]*100+[i for i in range(-7,500)]
|
||||||
|
f4.set_sequence_after_dct(sequence_after_dct)
|
||||||
|
info1=[0,1,0,1,1,0,1,0]
|
||||||
|
f4.write(info1)
|
||||||
|
# 读
|
||||||
|
sequence_after_dct2=f4.get_sequence_after_dct()
|
||||||
|
f4.set_sequence_after_dct(sequence_after_dct2)
|
||||||
|
info2=f4.read()
|
||||||
|
print (info2)
|
||||||
|
|
@ -0,0 +1,128 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Mon Jun 10 15:26:43 2019
|
||||||
|
|
||||||
|
@author: Administrator
|
||||||
|
"""
|
||||||
|
|
||||||
|
import math
|
||||||
|
|
||||||
|
class Jsteg:
|
||||||
|
def __init__(self):
|
||||||
|
self.sequence_after_dct=None
|
||||||
|
|
||||||
|
def set_sequence_after_dct(self,sequence_after_dct):
|
||||||
|
self.sequence_after_dct=sequence_after_dct
|
||||||
|
self.available_info_len=len([i for i in self.sequence_after_dct if i not in (-1,1,0)]) # 不是绝对可靠的
|
||||||
|
print ("Load>> 可嵌入",self.available_info_len,'bits')
|
||||||
|
|
||||||
|
def get_sequence_after_dct(self):
|
||||||
|
return self.sequence_after_dct
|
||||||
|
|
||||||
|
def write(self,info):
|
||||||
|
"""先嵌入信息的长度,然后嵌入信息"""
|
||||||
|
info=self._set_info_len(info)
|
||||||
|
info_len=len(info)
|
||||||
|
info_index=0
|
||||||
|
im_index=0
|
||||||
|
while True:
|
||||||
|
if info_index>=info_len:
|
||||||
|
break
|
||||||
|
data=info[info_index]
|
||||||
|
if self._write(im_index,data):
|
||||||
|
info_index+=1
|
||||||
|
im_index+=1
|
||||||
|
|
||||||
|
|
||||||
|
def read(self):
|
||||||
|
"""先读出信息的长度,然后读出信息"""
|
||||||
|
_len,sequence_index=self._get_info_len()
|
||||||
|
info=[]
|
||||||
|
info_index=0
|
||||||
|
|
||||||
|
while True:
|
||||||
|
if info_index>=_len:
|
||||||
|
break
|
||||||
|
data=self._read(sequence_index)
|
||||||
|
if data!=None:
|
||||||
|
info.append(data)
|
||||||
|
info_index+=1
|
||||||
|
sequence_index+=1
|
||||||
|
|
||||||
|
return info
|
||||||
|
|
||||||
|
#===============================================================#
|
||||||
|
|
||||||
|
def _set_info_len(self,info):
|
||||||
|
l=int(math.log(self.available_info_len,2))+1
|
||||||
|
info_len=[0]*l
|
||||||
|
_len=len(info)
|
||||||
|
info_len[-len(bin(_len))+2:]=[int(i) for i in bin(_len)[2:]]
|
||||||
|
return info_len+info
|
||||||
|
|
||||||
|
def _get_info_len(self):
|
||||||
|
l=int(math.log(self.available_info_len,2))+1
|
||||||
|
len_list=[]
|
||||||
|
_l_index=0
|
||||||
|
_seq_index=0
|
||||||
|
while True:
|
||||||
|
if _l_index>=l:
|
||||||
|
break
|
||||||
|
_d=self._read(_seq_index)
|
||||||
|
if _d!=None:
|
||||||
|
len_list.append(str(_d))
|
||||||
|
_l_index+=1
|
||||||
|
_seq_index+=1
|
||||||
|
_len=''.join(len_list)
|
||||||
|
_len=int(_len,2)
|
||||||
|
return _len,_seq_index
|
||||||
|
|
||||||
|
# 注意经过DCT会有负值,此处最低有效位的嵌入方式与空域LSB略有不同
|
||||||
|
def _write(self,index,data):
|
||||||
|
origin=self.sequence_after_dct[index]
|
||||||
|
if origin in (-1,1,0):
|
||||||
|
return False
|
||||||
|
|
||||||
|
lower_bit=origin%2
|
||||||
|
if lower_bit==data:
|
||||||
|
pass
|
||||||
|
elif origin>0:
|
||||||
|
if (lower_bit,data) == (0,1):
|
||||||
|
self.sequence_after_dct[index]=origin+1
|
||||||
|
elif (lower_bit,data) == (1,0):
|
||||||
|
self.sequence_after_dct[index]=origin-1
|
||||||
|
elif origin<0:
|
||||||
|
if (lower_bit,data) == (0,1):
|
||||||
|
self.sequence_after_dct[index]=origin-1
|
||||||
|
elif (lower_bit,data) == (1,0):
|
||||||
|
self.sequence_after_dct[index]=origin+1
|
||||||
|
|
||||||
|
return True
|
||||||
|
|
||||||
|
def _read(self,index):
|
||||||
|
if self.sequence_after_dct[index] not in (-1,1,0):
|
||||||
|
return self.sequence_after_dct[index]%2
|
||||||
|
else:
|
||||||
|
return None
|
||||||
|
'''
|
||||||
|
import cv2
|
||||||
|
import numpy as np
|
||||||
|
|
||||||
|
def dct(m):
|
||||||
|
m = np.float32(m)/255.0
|
||||||
|
return cv2.dct(m)*255
|
||||||
|
'''
|
||||||
|
|
||||||
|
if __name__=="__main__":
|
||||||
|
jsteg=Jsteg()
|
||||||
|
# 写
|
||||||
|
sequence_after_dct=[-1,0,1]*100+[i for i in range(-7,500)]
|
||||||
|
#print(sequence_after_dct)
|
||||||
|
jsteg.set_sequence_after_dct(sequence_after_dct)
|
||||||
|
info1=[0,1,0,1,1,0,1,0]
|
||||||
|
jsteg.write(info1)
|
||||||
|
sequence_after_dct2=jsteg.get_sequence_after_dct()
|
||||||
|
# 读
|
||||||
|
jsteg.set_sequence_after_dct(sequence_after_dct2)
|
||||||
|
info2=jsteg.read()
|
||||||
|
print (info2)
|
||||||
|
|
@ -0,0 +1 @@
|
||||||
|
dajkhfahjkf
|
||||||
|
|
@ -0,0 +1 @@
|
||||||
|
dajkhfahjkf
|
||||||
|
|
@ -0,0 +1,69 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Sun May 19 12:43:26 2019
|
||||||
|
|
||||||
|
@author: Administrator
|
||||||
|
"""
|
||||||
|
|
||||||
|
from PIL import Image
|
||||||
|
|
||||||
|
def mod(x,y):
|
||||||
|
return x%y
|
||||||
|
|
||||||
|
def toasc(strr):
|
||||||
|
return int(strr, 2)
|
||||||
|
|
||||||
|
|
||||||
|
#le为所要提取的信息的长度,str1为加密载体图片的路径,str2为提取文件的保存路径
|
||||||
|
def func(le,str1,str2):
|
||||||
|
b=""
|
||||||
|
im = Image.open(str1)
|
||||||
|
lenth = le*8
|
||||||
|
width,height = im.size[0],im.size[1]
|
||||||
|
count = 0
|
||||||
|
for h in range(height):
|
||||||
|
for w in range(width):
|
||||||
|
#获得(w,h)点像素的值
|
||||||
|
pixel = im.getpixel((w, h))
|
||||||
|
#此处余3,依次从R、G、B三个颜色通道获得最低位的隐藏信息
|
||||||
|
if count%3==0:
|
||||||
|
count+=1
|
||||||
|
b=b+str((mod(int(pixel[0]),2)))
|
||||||
|
if count ==lenth:
|
||||||
|
break
|
||||||
|
if count%3==1:
|
||||||
|
count+=1
|
||||||
|
b=b+str((mod(int(pixel[1]),2)))
|
||||||
|
if count ==lenth:
|
||||||
|
break
|
||||||
|
if count%3==2:
|
||||||
|
count+=1
|
||||||
|
b=b+str((mod(int(pixel[2]),2)))
|
||||||
|
if count ==lenth:
|
||||||
|
break
|
||||||
|
if count == lenth:
|
||||||
|
break
|
||||||
|
|
||||||
|
with open(str2,"w",encoding='utf-8') as f:
|
||||||
|
for i in range(0,len(b),8):
|
||||||
|
#以每8位为一组二进制,转换为十进制
|
||||||
|
stra = toasc(b[i:i+8])
|
||||||
|
#将转换后的十进制数视为ascii码,再转换为字符串写入到文件中
|
||||||
|
#print((stra))
|
||||||
|
f.write(chr(stra))
|
||||||
|
print("完成信息提取!")
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
def main():
|
||||||
|
#文件长度
|
||||||
|
le = 11
|
||||||
|
#含有隐藏信息的图片
|
||||||
|
new = "new.png"
|
||||||
|
#信息提取出后所存放的文件
|
||||||
|
get_info = "get_flag.txt"
|
||||||
|
func(le,new,get_info)
|
||||||
|
|
||||||
|
if __name__=='__main__':
|
||||||
|
main()
|
||||||
|
|
@ -0,0 +1,85 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Sun May 19 11:20:05 2019
|
||||||
|
|
||||||
|
@author: Administrator
|
||||||
|
"""
|
||||||
|
|
||||||
|
from PIL import Image
|
||||||
|
|
||||||
|
|
||||||
|
def plus(string):
|
||||||
|
#Python zfill() 方法返回指定长度的字符串,原字符串右对齐,前面填充0。
|
||||||
|
return string.zfill(8)
|
||||||
|
|
||||||
|
def get_key(strr):
|
||||||
|
#获取要隐藏的文件内容
|
||||||
|
with open(strr,"rb") as f:
|
||||||
|
s = f.read()
|
||||||
|
string=""
|
||||||
|
for i in range(len(s)):
|
||||||
|
#逐个字节将要隐藏的文件内容转换为二进制,并拼接起来
|
||||||
|
#1.先用ord()函数将s的内容逐个转换为ascii码
|
||||||
|
#2.使用bin()函数将十进制的ascii码转换为二进制
|
||||||
|
#3.由于bin()函数转换二进制后,二进制字符串的前面会有"0b"来表示这个字符串是二进制形式,所以用replace()替换为空
|
||||||
|
#4.又由于ascii码转换二进制后是七位,而正常情况下每个字符由8位二进制组成,所以使用自定义函数plus将其填充为8位
|
||||||
|
string=string+""+plus(bin(s[i]).replace('0b',''))
|
||||||
|
#print(string)
|
||||||
|
return string
|
||||||
|
|
||||||
|
def mod(x,y):
|
||||||
|
return x%y
|
||||||
|
|
||||||
|
#str1为载体图片路径,str2为隐写文件,str3为加密图片保存的路径
|
||||||
|
def func(str1,str2,str3):
|
||||||
|
im = Image.open(str1)
|
||||||
|
#获取图片的宽和高
|
||||||
|
width,height= im.size[0],im.size[1]
|
||||||
|
print("width:"+str(width))
|
||||||
|
print("height:"+str(height))
|
||||||
|
count = 0
|
||||||
|
#获取需要隐藏的信息
|
||||||
|
key = get_key(str2)
|
||||||
|
keylen = len(key)
|
||||||
|
for h in range(height):
|
||||||
|
for w in range(width):
|
||||||
|
pixel = im.getpixel((w,h))
|
||||||
|
a=pixel[0]
|
||||||
|
b=pixel[1]
|
||||||
|
c=pixel[2]
|
||||||
|
if count == keylen:
|
||||||
|
break
|
||||||
|
#下面的操作是将信息隐藏进去
|
||||||
|
#分别将每个像素点的RGB值余2,这样可以去掉最低位的值
|
||||||
|
#再从需要隐藏的信息中取出一位,转换为整型
|
||||||
|
#两值相加,就把信息隐藏起来了
|
||||||
|
a= a-mod(a,2)+int(key[count])
|
||||||
|
count+=1
|
||||||
|
if count == keylen:
|
||||||
|
im.putpixel((w,h),(a,b,c))
|
||||||
|
break
|
||||||
|
b =b-mod(b,2)+int(key[count])
|
||||||
|
count+=1
|
||||||
|
if count == keylen:
|
||||||
|
im.putpixel((w,h),(a,b,c))
|
||||||
|
break
|
||||||
|
c= c-mod(c,2)+int(key[count])
|
||||||
|
count+=1
|
||||||
|
if count == keylen:
|
||||||
|
im.putpixel((w,h),(a,b,c))
|
||||||
|
break
|
||||||
|
if count % 3 == 0:
|
||||||
|
im.putpixel((w,h),(a,b,c))
|
||||||
|
im.save(str3)
|
||||||
|
|
||||||
|
def main():
|
||||||
|
#原图
|
||||||
|
old = "old.png"
|
||||||
|
#处理后输出的图片路径
|
||||||
|
new = "new.png"
|
||||||
|
#需要隐藏的信息
|
||||||
|
enc = "flag.txt"
|
||||||
|
func(old,enc,new)
|
||||||
|
|
||||||
|
if __name__=='__main__':
|
||||||
|
main()
|
||||||
Binary file not shown.
|
After Width: | Height: | Size: 160 KiB |
Binary file not shown.
|
After Width: | Height: | Size: 161 KiB |
|
|
@ -0,0 +1,282 @@
|
||||||
|
# PDF格式学习
|
||||||
|
## PDF简介
|
||||||
|
* PDF是Portable Document Format 的缩写,可翻译为“便携文件格式”,由Adobe System Incorporated 公司在1992年发明。
|
||||||
|
|
||||||
|
* PDF文件是一种编程形式的文档格式,它所有显示的内容,都是通过相应的操作符进行绘制的。
|
||||||
|
* PDF基本显示单元包括:文字,图片,矢量图,图片
|
||||||
|
* PDF扩展单元包括:水印,电子署名,注释,表单,多媒体,3D
|
||||||
|
* PDF动作单元:书签,超链接(拥有动作的单元有很多个,包括电子署名,多媒体等等)
|
||||||
|
## PDF的优点
|
||||||
|
* 一致性:
|
||||||
|
在所有可以打开PDF的机器上,展示的效果是完全一致,不会出现段落错乱、文字乱码这些排版问题。尤其是文档中,本身可以嵌入字体,避免了客户端没有对应字体,而导致文字显示不一致的问题。所以,在印刷行业,绝大多数用的都是PDF格式。
|
||||||
|
* 不易修改:
|
||||||
|
用过PDF文件的人,都会知道,对已经保存之后的PDF文件,想要进行重新排版,基本上就不可能的,这就保证了从资料源发往外界的资料,不容易被篡改。
|
||||||
|
* 安全性:
|
||||||
|
PDF文档可以进行加密,包括以下几种加密形式:文档打开密码,文档权限密码,文档证书密码,加密的方法包括:RC4,AES,通过加密这种形式,可以达到资料防扩散等目的。
|
||||||
|
* 不失真:
|
||||||
|
PDF文件中,使用了矢量图,在文件浏览时,无论放大多少倍,都不会导致使用矢量图绘制的文字,图案的失真。
|
||||||
|
* 支持多种压缩方式:
|
||||||
|
为了减少PDF文件的size,PDF格式支持各种压缩方式: asciihex,ascii85,lzw,runlength,ccitt,jbig2,jpeg(DCT),jpeg2000(jpx)
|
||||||
|
* 支持多种印刷标准:
|
||||||
|
支持PDF-A,PDF-X
|
||||||
|
|
||||||
|
## PDF格式
|
||||||
|
根据PDF官方指南,理解PDF格式可以从四个方面下手——**Objects**(对象)、**File structure**(物理文件结构)、**Document structure**(逻辑文件结构)、**Content streams**(内容流)。
|
||||||
|
|
||||||
|
### 对象
|
||||||
|
|
||||||
|
### 物理文件结构
|
||||||
|
* 整体上分为文件头(Header)、对象集合(Body)、交叉引用表(Xref table)、文件尾(Trailer)四个部分,结构如图。修改过的PDF结构会有部分变化。
|
||||||
|
* 未经修改
|
||||||
|

|
||||||
|
* 经修改
|
||||||
|
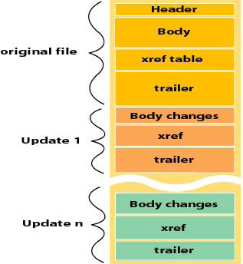
|
||||||
|
#### 文件头
|
||||||
|
* 文件头是PDF文件的第一行,格式如下:
|
||||||
|
```
|
||||||
|
%PDF-1.7
|
||||||
|
```
|
||||||
|
* 这是个固定格式,表示这个PDF文件遵循的PDF规范版本,解析PDF的时候尽量支持高版本的规范,以保证支持大多数工具生成的PDF文件。1.7版本支持1.0-1.7之间的所有版本。
|
||||||
|
|
||||||
|
#### 对象集合
|
||||||
|
* 这是一个PDF文件最重要的部分,文件中用到的所有对象,包括文本、图象、音乐、视频、字体、超连接、加密信息、文档结构信息等等,都在这里定义。格式如下:
|
||||||
|
```
|
||||||
|
2 0 obj
|
||||||
|
...
|
||||||
|
end obj
|
||||||
|
```
|
||||||
|
* 一个对象的定义包含4个部分:前面的2是**对象序号**,其用来唯一标记一个对象;0是**生成号**,按照PDF规范,如果一个PDF文件被修改,那这个数字是累加的,它和对象序号一起标记是原始对象还是修改后的对象,但是实际开发中,很少有用这种方式修改PDF的,都是重新编排对象号;obj和endobj是对象的定义范围,可以抽象的理解为这就是一个左括号和右括号;省略号部分是PDF规定的任意合法对象。
|
||||||
|
* 可以通过R关键字来引用任何一个对象,比如要引用上面的对象,可以使用2 0 R,需要主意的是,R关键字不仅可以引用一个已经定义的对象,还可以引用一个并**不存在的对象**,而且效果就和引用了一个空对象一样。
|
||||||
|
* 对象主要有下面几种
|
||||||
|
* **booleam**
|
||||||
|
用关键字true或false表示,可以是array对象的一个元素,或dictionary对象的一个条目。也可以用在PostScript计算函数里面,做为if或if esle的一个条件。
|
||||||
|
* **numeric**
|
||||||
|
包括整形和实型,不支持非十进制数字,不支持指数形式的数字。
|
||||||
|
例:
|
||||||
|
1)整数 123 4567 +111 -2
|
||||||
|
范围:正2的31次方-1到负的2的31次方
|
||||||
|
2)实数 12.3 0.8 +6.3 -4.01 -3. +.03
|
||||||
|
范围:±3.403 ×10的38次方 ±1.175 × 10的-38次方
|
||||||
|
* 注意:如果整数超过表示范围将转化成实数,如果实数超过范围就会出错
|
||||||
|
* **string**
|
||||||
|
由一系列0-255之间的字节组成,一个string总长度不能超过65535.string有以下两种方式:
|
||||||
|
* 十六进制字串
|
||||||
|
由<>包含起来的一个16进制串,两位表示一个字符,不足两位用0补齐。
|
||||||
|
例: \<Aabb> 表示AA和BB两个字符 \<AAB> 表示AA和B0两个字符
|
||||||
|
* 直接字串
|
||||||
|
由()包含起来的一个字串,中间可以使用转义符"/"。
|
||||||
|
例:
|
||||||
|
(abc) 表示abc
|
||||||
|
(a//) 表示a/
|
||||||
|
转义符的定义如下:
|
||||||
|
|
||||||
|
|转义字符| 含义|
|
||||||
|
|--------|--------|
|
||||||
|
|/n |换行|
|
||||||
|
/r |回车
|
||||||
|
/t |水平制表符
|
||||||
|
/b |退格
|
||||||
|
/f |换页(Form feed (FF))
|
||||||
|
/( |左括号
|
||||||
|
/) |右括号
|
||||||
|
// |反斜杠
|
||||||
|
/ddd |八进制形式的字符
|
||||||
|
|
||||||
|
|
||||||
|
* 对象类别(续)
|
||||||
|
|
||||||
|
* **name**
|
||||||
|
由一个前导/和后面一系列字符组成,最大长度为127。和string不同的是,name是**不可分割**的并且是**唯一**的,不可分割就是说一个name对象就是一个原子,比如/name,不能说n就是这个name的一个元素;唯一就是指两个相同的name一定代表同一个对象。从pdf1.2开始,除了ascii的0,别的都可以用一个#加两个十六进制的数字表示。
|
||||||
|
例:
|
||||||
|
/name 表示name
|
||||||
|
/name#20is 表示name is
|
||||||
|
/name#200 表示name 0
|
||||||
|
* **array**
|
||||||
|
用[]包含的一组对象,可以是任何pdf对象(包括array)。虽然pdf只支持一维array,但可以通过array的嵌套实现任意维数的array(但是一个array的元素不能超过8191)。
|
||||||
|
例:[549 3.14 false (Ralph) /SomeName]
|
||||||
|
* **Dictionary**
|
||||||
|
用"<<"和">>"包含的若干组条目,每组条目都由key和value组成,其中key必须是name对象,并且一个dictionary内的key是唯一的;value可以是任何pdf的合法对象(包括dictionary对象)。
|
||||||
|
例:
|
||||||
|
```
|
||||||
|
<< /IntegerItem 12
|
||||||
|
/StringItem (a string)
|
||||||
|
/Subdictionary
|
||||||
|
<< /Item1 0.4
|
||||||
|
/Item2 true
|
||||||
|
/LastItem (not!)
|
||||||
|
/VeryLastItem (OK)
|
||||||
|
>>
|
||||||
|
>>
|
||||||
|
```
|
||||||
|
* **stream**
|
||||||
|
由一个字典和紧跟其后面的一组关键字stream和endstream以及这组关键字中间包含一系列字节组成。内容和string很相似,但有区别:stream可以分几次读取,分开使用不同的部分,string必须作为一个整体一次全部读取使用;string有长度限制,但stream却没有这个限制。一般较大的数据都用stream表示。需要注意的是,stream必须是间接对象,并且stream的字典必须是直接对象。从1.2规范以后,stream可以以外部文件形式存在,这种情况下,解析PDF的时候stream和endstream之间的内容就被忽略掉。
|
||||||
|
例:
|
||||||
|
```
|
||||||
|
dictionary
|
||||||
|
stream
|
||||||
|
…data…
|
||||||
|
endstream
|
||||||
|
```
|
||||||
|
stream字典中常用的字段如下:
|
||||||
|
|
||||||
|
|字段名 |类型| 值|
|
||||||
|
|--------|--------|--------|
|
||||||
|
|Length| 整形|(必须)关键字stream和endstream之间的数据长度,endstream之前可能会有一个多余的EOL标记,这个不计算在数据的长度中。
|
||||||
|
Filter |名字 或 数组 |(可选)Stream的编码算法名称(列表)。如果有多个,则数组中的编码算法列表顺序就是数据被编码的顺序。
|
||||||
|
DecodeParms |字典 或 数组 |(可选)一个参数字典或由参数字典组成的一个数组,供Filter使用。如果仅有一个Filter并且这个Filter需要参数,除非这个Filter的所有参数都已经给了默认值,否则的话 DecodeParms必须设置给Filter。如果有多个Filter,并且任意一个Filter使用了非默认的参数, DecodeParms 必须是个数组,每个元素对应一个Filter的参数列表(如果某个Filter无需参数或所有参数都有了默认值,就用空对象代替)。 如果没有Filter需要参数,或者所有Filter的参数都有默认值,DecodeParms 就被忽略了。
|
||||||
|
F |文件标识 |(可选)保存stream数据的文件。如果有这个字段, stream和endstream就被忽略,FFilter将会代替Filter, FDecodeParms将代替DecodeParms。Length字段还是表示stream和endstream之间数据的长度,但是通常此刻已经没有数据了,长度是0.
|
||||||
|
FFilter |名字 或 字典| (可选)和filter类似,针对外部文件。
|
||||||
|
FDecodeParms |字典 或 数组| (可选)和DecodeParams类似,针对外部文件。
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
Stream的编码算法名称(列表)。如果有多个,则数组中的编码算法列表顺序就是数据被编码的顺序。且需要被编码。编码算法主要如下:
|
||||||
|
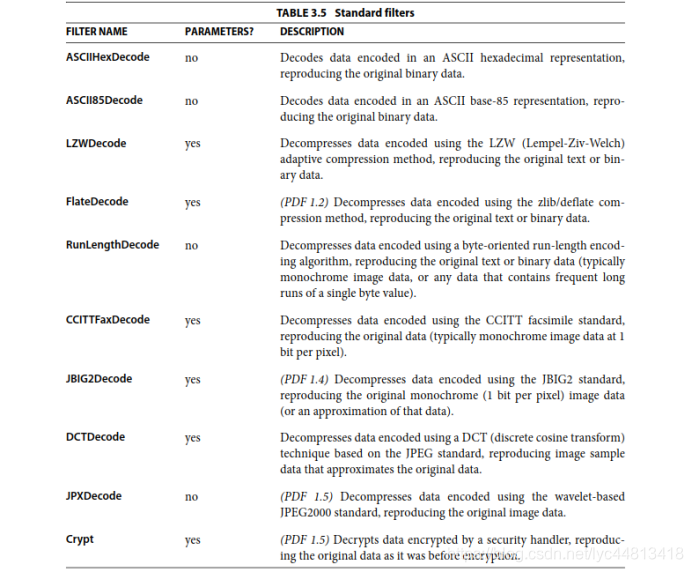
|
||||||
|
编码可视化主要显示为乱码,所以提供了隐藏信息的机会,如下图的steam内容为乱码。
|
||||||
|

|
||||||
|
* **NULL**
|
||||||
|
用null表示,代表空。如果一个key的值为null,则这个key可以被忽略;如果引用一个不存在的object则等价于引用一个空对象。
|
||||||
|
|
||||||
|
#### 交叉引用表
|
||||||
|
* 交叉引用表是PDf文件内部一种特殊的文件组织方式,可以很方便的根据对象号随机访问一个对象。其格式如下:
|
||||||
|
```
|
||||||
|
xref
|
||||||
|
0 1
|
||||||
|
0000000000 65535 f
|
||||||
|
4 1
|
||||||
|
0000000009 00000 n
|
||||||
|
8 3
|
||||||
|
0000000074 00000 n
|
||||||
|
0000000120 00000 n
|
||||||
|
0000000179 00000 n
|
||||||
|
```
|
||||||
|
* 其中,xref是开始标志,表示以下为一个交叉引用表的内容;每个交叉引用表又可以分为若干个子段,每个子段的第一行是两个数字,第一个是对象起始号,后面是连续的对象个数,接着每行是这个子段的每个对象的具体信息——每行的前10个数字代表这个这个对象**相对文件头的偏移地址**,后面的5位数字是**生成号**(用于标记PDF的更新信息,和对象的生成号作用类似),最后一位f或n表示对象是否被使用(n表示使用,f表示被删除或没有用)。上面这个交叉引用表一共有3个子段,分别有1个,1个,3个对象,第一个子段的对象不可用,其余子段对象可用。
|
||||||
|
#### 文件尾
|
||||||
|
* 通过trailer可以快速的找到交叉引用表的位置,进而可以精确定位每一个对象;还可以通过它本身的字典还可以获取文件的一些全局信息(作者,关键字,标题等),加密信息,等等。具体形式如下:
|
||||||
|
```
|
||||||
|
trailer
|
||||||
|
<<
|
||||||
|
key1 value1
|
||||||
|
key2 value2
|
||||||
|
key3 value3
|
||||||
|
…
|
||||||
|
>>
|
||||||
|
startxref
|
||||||
|
553
|
||||||
|
%%EOF
|
||||||
|
```
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
* trailer后面紧跟一个字典,包含若干键-值对。具体含义如下:
|
||||||
|
|
||||||
|
|键 |值类型| 值说明|
|
||||||
|
|--------|--------|--------|
|
||||||
|
|Size| 整形数字| 所有间接对象的个数。一个PDF文件,如果被更新过,则会有多个对象集合、交叉引用表、trailer,最后一个trailer的这个字段记录了之前所有对象的个数。这个值必须是直接对象。|
|
||||||
|
|Prev |整形数字| 当文件有多个对象集合、交叉引用表和trailer时,才会有这个键,它表示前一个相对于文件头的偏移位置。这个值必须是直接对象。|
|
||||||
|
|Root |字典 |Catalog字典(文件的逻辑入口点)的对象号。必须是间接对象。|
|
||||||
|
|Encrypt |字典| 文档被保护时,会有这个字段,加密字典的对象号。|
|
||||||
|
|Info |字典 |存放文档信息的字典,必须是间接对象。|
|
||||||
|
|ID |数组 |文件的ID|
|
||||||
|
|
||||||
|
* 上面代码中的startxref:后面的数字表示最后一个交叉引用表相对于文件起始位置的偏移量
|
||||||
|
* %%EOF:文件结束符
|
||||||
|
### 逻辑文件结构
|
||||||
|
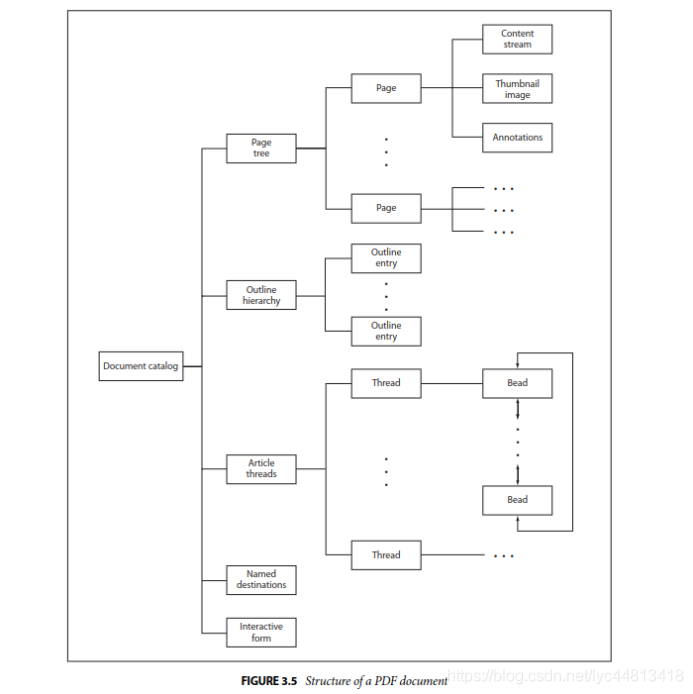
|
||||||
|
|
||||||
|
#### catalog根节点
|
||||||
|
* catalog是整个PDF逻辑结构的根节点,这个可以通过trailer的Root字段定位,虽然简单,但是相当重要,因为这里是PDF文件物理结构和逻辑结构的连接点。Catalog字典包含的信息非常多,这里仅就最主要的几个字段做个说明。
|
||||||
|
* Pages字段
|
||||||
|
这是个必须字段,是PDF里面所有页面的描述集合。Pages字段本身是个字典,它里面又包含了一下几个主要字段:
|
||||||
|
|
||||||
|
|字段 |类型 |值|
|
||||||
|
|--------|--------|--------|
|
||||||
|
Type |name| (必须)只能为Pages 。
|
||||||
|
Parent |dictionary |(如果不是catalog里面指定的跟节点,则必须有,并且必须是间接对象) 当前节点的直接父节点。
|
||||||
|
Kids |array |(必须)一个间接对象组成的数组,节点可能是page或page tree。
|
||||||
|
Count |integer| (必须) page tree里面所包含叶子节点(page 对象)的个数。
|
||||||
|
|
||||||
|
从以上字段可以看出,Pages最主要的功能就是组织所有的page对象。Page对象描述了一个PDF页面的属性、资源等信息。Page对象是一个字典,它主要包含一下几个重要的属性:
|
||||||
|
|
||||||
|
|字段 |类型 |值|
|
||||||
|
|--------|--------|--------|
|
||||||
|
Type |name |(必须)必须是Page。
|
||||||
|
Parent |dictionary| (必须;并且只能是间接对象)当前page节点的直接父节点page tree 。
|
||||||
|
LastModified| date| (如果存在PieceInfo字段,就必须有,否则可选)记录当前页面被最后一次修改的日期和时间。
|
||||||
|
Resources| dictionary| (必须; 可继承)记录了当前page用到的所有资源。如果当前页不用任何资源,则这是个空字典。忽略所有字段则表示继承父节点的资源。
|
||||||
|
MediaBox |rectangle| (必须; 可继承)定义了要显示或打印页面的物理媒介的区域(default user space units)
|
||||||
|
CropBox |rectangle| (可选; 可继承)定义了一个可视区域,当前页被显示或打印的时候,它的内容会被这个区域裁剪。默认值就是 MediaBox。
|
||||||
|
BleedBox|rectangle |(可选) 定义了一个区域,当输出设备是个生产环境( production environment)的时候,页面显示的内容会被裁剪。默认值是 CropBox.
|
||||||
|
Contents |stream or array| (可选) 描述页面内容的流。如果这个字段缺省,则页面上什么也不会显示。这个值可以是一个流,也可以是由几个流组成的一个数组。如果是数组,实际效果相当于所有的流是按顺序连在一起的一个流,这就允许PDF生成的时候可以随时插入图片或其他资源。流之间的分割只是词汇上的一个分割,并不是逻辑上或者组织形式的切割。
|
||||||
|
Rotate |integer| (可选; 可继承) 顺时钟旋转的角度数,这个必须是90的整数倍,默认是0。
|
||||||
|
Thumb| stream |(可选)定义当前页的缩略图。
|
||||||
|
Annots| array| (可选) 和当前页面关联的注释。
|
||||||
|
Metadata |stream| (可选) 当前页包含的元数据。
|
||||||
|
一个简单例子:
|
||||||
|
```
|
||||||
|
3 0 obj
|
||||||
|
<< /Type /Page
|
||||||
|
/Parent 4 0 R
|
||||||
|
/MediaBox [ 0 0 612 792 ]
|
||||||
|
/Resources <</Font<<
|
||||||
|
/F3 7 0 R /F5 9 0 R /F7 11 0 R
|
||||||
|
>>
|
||||||
|
/ProcSet [ /PDF ]
|
||||||
|
>>
|
||||||
|
/Contents 12 0 R
|
||||||
|
/Thumb 14 0 R
|
||||||
|
/Annots [ 23 0 R 24 0 R]
|
||||||
|
>>
|
||||||
|
endobj
|
||||||
|
```
|
||||||
|
* Outlines字段
|
||||||
|
Outline是PDF里面为了方便用户从PDF的一部分跳转到另外一部分而设计的,有时候也叫书签(Bookmark),它是一个树状结构,可以直观的把PDF文件结构展现给用户。用户可以通过鼠标点击来打开或者关闭某个outline项来实现交互,当打开一个outline时,用户可以看到它的所有子节点,关闭一个outline的时候,这个outline的所有子节点会自动隐藏。并且,在点击的时候,阅读器会自动跳转到outline对应的页面位置。Outlines包含以下几个字段:
|
||||||
|
|
||||||
|
|字段 |类型 |值|
|
||||||
|
|--------|--------|--------|
|
||||||
|
Type |name |(可选)如果这个字段有值,则必须是Outlines。
|
||||||
|
First |dictionary |(必须;必须是间接对象) 第一个顶层Outline item。
|
||||||
|
Last |dictionary |(必须;必须是间接对象)最后一个顶层outline item。
|
||||||
|
Count |integer |(必须)outline的所有层次的item的总数。
|
||||||
|
|
||||||
|
Outline是一个管理outline item的顶层对象,我们看到的,其实是outline item,这个里面才包含了文字、行为、目标区域等等。一个outline item主要有一下几个字段:
|
||||||
|
|
||||||
|
|字段 |类型 |值|
|
||||||
|
|--------|--------|--------|
|
||||||
|
Title |text string| (必须)当前item要显示的标题。
|
||||||
|
Parent |dictionary |(必须;必须是间接对象) outline层级中,当前item的父对象。如果item本身是顶级item,则父对象就是它本身。
|
||||||
|
Prev| dictionary| (除了每层的第一个item外,其他item必须有这个字段;必须是间接对象)当前层级中,此item的前一个item。
|
||||||
|
Next |dictionary| (除了每层的最后一个item外,其他item必须有这个字段;必须是间接对象)当前层级中,此item的后一个item。
|
||||||
|
First |dictionary| (如果当前item有任何子节点,则这个字段是必须的;必须是间接对象) 当前item的第一个直接子节点。
|
||||||
|
Last |dictionary| (如果当前item有任何子节点,则这个字段是必须的;必须是间接对象) 当前item的最后一个直接子节点。
|
||||||
|
Dest |name,byte string, or array |(可选; 如果A字段存在,则这个不能被会略)当前的outline item被激活的时候,要显示的区域。
|
||||||
|
A |dictionary| (可选; 如果Dest 字段存在,则这个不能被忽略)当前的outline item被激活的时候,要执行的动作。
|
||||||
|
|
||||||
|
* URI字段
|
||||||
|
URI(uniform resource identifier),定义了文档级别的统一资源标识符和相关链接信息。目录和文档中的链接就是通过这个字段来处理的.
|
||||||
|
* Metadata字段
|
||||||
|
文档的一些附带信息,用xml表示,符合adobe的xmp规范。这个可以方便程序不用解析整个文件就能获得文件的大致信息。
|
||||||
|
* 其他
|
||||||
|
Catalog字典中,常用的字段一般有以下一些:
|
||||||
|
|
||||||
|
|字段 |类型 |值|
|
||||||
|
|--------|--------|--------|
|
||||||
|
Type |name| (必须)必须为Catalog。
|
||||||
|
Version| name |(可选)PDF文件所遵循的版本号(如果比文件头指定的版本号高的话)。如果这个字段缺省或者文件头指定的版本比这里的高,那就以文件头为准。一个PDF生成程序可以通过更新这个字段的值来修改PDF文件版本号。
|
||||||
|
Pages |dictionary| (必须并且必须为间接对象)当前文档的页面集合入口。
|
||||||
|
PageLabels |number tree| (可选) number tree,定义了页面和页面label对应关系。
|
||||||
|
Names| dictionary| (可选)文档的name字典。
|
||||||
|
Dests |dictionary |(可选;必须是间接对象)name和相应目标对应关系字典。
|
||||||
|
ViewerPreferences| dictionary| (可选)阅读参数配置字典,定义了文档被打开时候的行为。如果缺省,则使用阅读器自己的配置。
|
||||||
|
PageLayout |name |(可选) 指定文档被打开的时候页面的布局方式。SinglePageDisplay 单页OneColumnDisplay 单列TwoColumnLeftDisplay 双列,奇数页在左TwoColumnRightDisplay 双列,奇数页在右TwoPageLeft 双页,奇数页在左TwoPageRight 双页,奇数页在右缺省值: SinglePage.
|
||||||
|
PageMode |name| (可选) 当文档被打开时,指定文档怎么显示UseNone 目录和缩略图都不显示UseOutlines 显示目录UseThumbs 显示缩略图FullScreen 全屏模式,没有菜单,任何其他窗口UseOC 显示Optional content group 面板UseAttachments显示附件面板缺省值: UseNone.
|
||||||
|
Outlines |dictionary| (可选;必须为间接对象)文档的目录字典
|
||||||
|
Threads |array |(可选;必须为间接对象)文章线索字典组成的数组。
|
||||||
|
OpenAction |array or dictionary| (可选) 指定一个区域或一个action,在文档打开的时候显示(区域)或者执行(action)。如果缺省,则会用默认缩放率显示第一页的顶部。
|
||||||
|
AA |dictionary| (可选)一个附加的动作字典,在全局范围内定义了响应各种事件的action。
|
||||||
|
URI| dictionary |(可选)一个URI字典包含了文档级别的URI action信息。
|
||||||
|
AcroForm| dictionary |(可选)文档的交互式form (AcroForm)字典。
|
||||||
|
Metadata |stream |(可选;必须是间接对象)文档包含的元数据流。
|
||||||
|
|
||||||
|
|
||||||
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
File diff suppressed because it is too large
Load Diff
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
File diff suppressed because it is too large
Load Diff
Binary file not shown.
Binary file not shown.
Binary file not shown.
|
|
@ -0,0 +1,183 @@
|
||||||
|
# imports
|
||||||
|
import json
|
||||||
|
import time
|
||||||
|
import pickle
|
||||||
|
import scipy.misc
|
||||||
|
import skimage.io
|
||||||
|
import caffe
|
||||||
|
|
||||||
|
import numpy as np
|
||||||
|
import os.path as osp
|
||||||
|
|
||||||
|
from random import shuffle
|
||||||
|
#from PIL import Image
|
||||||
|
|
||||||
|
import matplotlib.image as mpimg
|
||||||
|
|
||||||
|
|
||||||
|
class AugmentDataLayerSync(caffe.Layer):
|
||||||
|
|
||||||
|
"""
|
||||||
|
This is a simple syncronous datalayer for inputting the augmented data layer on the fly
|
||||||
|
"""
|
||||||
|
|
||||||
|
def setup(self, bottom, top):
|
||||||
|
|
||||||
|
self.top_names = ['data', 'label']
|
||||||
|
|
||||||
|
# === Read input parameters ===
|
||||||
|
|
||||||
|
# params is a python dictionary with layer parameters.
|
||||||
|
params = eval(self.param_str)
|
||||||
|
|
||||||
|
# Check the paramameters for validity.
|
||||||
|
check_params(params)
|
||||||
|
|
||||||
|
# store input as class variables
|
||||||
|
self.batch_size = params['batch_size']
|
||||||
|
|
||||||
|
# Create a batch loader to load the images.
|
||||||
|
self.batch_loader = BatchLoader( params, None )
|
||||||
|
|
||||||
|
# === reshape tops ===
|
||||||
|
# since we use a fixed input image size, we can shape the data layer
|
||||||
|
# once. Else, we'd have to do it in the reshape call.
|
||||||
|
top[0].reshape( self.batch_size,
|
||||||
|
1,
|
||||||
|
params['im_shape'][0],
|
||||||
|
params['im_shape'][1] )
|
||||||
|
|
||||||
|
# Ground truth
|
||||||
|
top[1].reshape(self.batch_size)
|
||||||
|
|
||||||
|
print_info( "AugmentStegoDataLayerSync", params )
|
||||||
|
|
||||||
|
def forward(self, bottom, top):
|
||||||
|
"""
|
||||||
|
Load data.
|
||||||
|
"""
|
||||||
|
for itt in range(self.batch_size):
|
||||||
|
# Use the batch loader to load the next image.
|
||||||
|
im, label = self.batch_loader.load_next_image()
|
||||||
|
|
||||||
|
# Add directly to the caffe data layer
|
||||||
|
top[0].data[itt, 0, :, :] = im
|
||||||
|
top[1].data[itt] = label
|
||||||
|
|
||||||
|
def reshape(self, bottom, top):
|
||||||
|
"""
|
||||||
|
There is no need to reshape the data, since the input is of fixed size
|
||||||
|
(rows and columns)
|
||||||
|
"""
|
||||||
|
pass
|
||||||
|
|
||||||
|
def backward(self, top, propagate_down, bottom):
|
||||||
|
"""
|
||||||
|
These layers does not back propagate
|
||||||
|
"""
|
||||||
|
pass
|
||||||
|
|
||||||
|
|
||||||
|
class BatchLoader(object):
|
||||||
|
|
||||||
|
"""
|
||||||
|
This class abstracts away the loading of images.
|
||||||
|
Images can either be loaded singly, or in a batch. The latter is used for
|
||||||
|
the asyncronous data layer to preload batches while other processing is
|
||||||
|
performed.
|
||||||
|
"""
|
||||||
|
|
||||||
|
def __init__(self, params, result):
|
||||||
|
|
||||||
|
self.result = result
|
||||||
|
self.batch_size = params['batch_size']
|
||||||
|
self.root = params['root']
|
||||||
|
self.im_shape = params['im_shape']
|
||||||
|
self.trainMode = ( params['split'] == 'train' ) # determine the mode, if test, no augment
|
||||||
|
|
||||||
|
# get list of image indexes.
|
||||||
|
list_file = params['split'] + '.txt'
|
||||||
|
TXT_FILE = osp.join( self.root, list_file )
|
||||||
|
txt_lines = [ line.rstrip('\n') for line in open( TXT_FILE ) ]
|
||||||
|
|
||||||
|
total_size = len( txt_lines )
|
||||||
|
|
||||||
|
assert total_size%2 == 0, "total_size must be even"
|
||||||
|
|
||||||
|
self.images = []
|
||||||
|
self.labels = np.zeros( ( total_size, ), dtype = np.int64 )
|
||||||
|
self.indexlist = range( total_size )
|
||||||
|
|
||||||
|
for i in np.arange(total_size):
|
||||||
|
tmp = txt_lines[i].split()
|
||||||
|
self.images.append(tmp[0])
|
||||||
|
self.labels[i] = int(tmp[1])
|
||||||
|
|
||||||
|
self._cur = 0 # current image
|
||||||
|
self._epoch = 0 # current epoch count, also used as the randomization seed
|
||||||
|
self._flp = 1 # Augment flip number,
|
||||||
|
self._rot = 0 # Augment rotation number
|
||||||
|
|
||||||
|
print "BatchLoader initialized with {} images".format(len(self.indexlist))
|
||||||
|
|
||||||
|
def load_next_image( self ):
|
||||||
|
|
||||||
|
"""
|
||||||
|
Load the next image in a batch
|
||||||
|
"""
|
||||||
|
# Did we finish an epoch
|
||||||
|
if self._cur == len(self.indexlist):
|
||||||
|
self._epoch += 1
|
||||||
|
l = np.random.seed( self._epoch ) #randomize, aslo reproducible
|
||||||
|
l = np.random.permutation( len(self.indexlist)/2 )
|
||||||
|
l2 = np.vstack( ( 2*l, 2*l + 1 )).T
|
||||||
|
self.indexlist = l2.reshape(len(self.indexlist),)
|
||||||
|
self._cur = 0
|
||||||
|
|
||||||
|
# Index list
|
||||||
|
index = self.indexlist[self._cur]
|
||||||
|
|
||||||
|
#load an image
|
||||||
|
image_file_name = self.images[index]
|
||||||
|
|
||||||
|
im = np.asarray( mpimg.imread( image_file_name ))
|
||||||
|
|
||||||
|
#Determine the new fliplr and rot90 status, used it in the stego
|
||||||
|
if ( self.trainMode ):
|
||||||
|
if ( self._cur % 2 == 0 ):
|
||||||
|
self._flp = np.random.choice(2)*2 - 1
|
||||||
|
self._rot = np.random.randint(4)
|
||||||
|
im = im[:,::self._flp]
|
||||||
|
im = np.rot90(im, self._rot)
|
||||||
|
|
||||||
|
#load the ground truth
|
||||||
|
label = self.labels[index]
|
||||||
|
|
||||||
|
self._cur += 1
|
||||||
|
|
||||||
|
return im, label
|
||||||
|
|
||||||
|
|
||||||
|
def check_params(params):
|
||||||
|
"""
|
||||||
|
A utility function to check the parameters for the data layers.
|
||||||
|
"""
|
||||||
|
assert 'split' in params.keys(
|
||||||
|
), 'Params must include split (train, val, or test).'
|
||||||
|
|
||||||
|
required = ['batch_size', 'root', 'im_shape']
|
||||||
|
for r in required:
|
||||||
|
assert r in params.keys(), 'Params must include {}'.format(r)
|
||||||
|
|
||||||
|
|
||||||
|
def print_info(name, params):
|
||||||
|
"""
|
||||||
|
Ouput some info regarding the class
|
||||||
|
"""
|
||||||
|
print "{} initialized for split: {}, with bs: {}, im_shape: {}.".format(
|
||||||
|
name,
|
||||||
|
params['split'],
|
||||||
|
params['batch_size'],
|
||||||
|
params['im_shape'])
|
||||||
|
|
||||||
|
|
||||||
|
|
@ -0,0 +1,122 @@
|
||||||
|
{
|
||||||
|
"cells": [
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": 1,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": false,
|
||||||
|
"deletable": true,
|
||||||
|
"editable": true
|
||||||
|
},
|
||||||
|
"outputs": [
|
||||||
|
{
|
||||||
|
"name": "stdout",
|
||||||
|
"output_type": "stream",
|
||||||
|
"text": [
|
||||||
|
"[HG:] Train Set = 30000, Test = 10000\n"
|
||||||
|
]
|
||||||
|
}
|
||||||
|
],
|
||||||
|
"source": [
|
||||||
|
"import numpy as np\n",
|
||||||
|
"import os\n",
|
||||||
|
"\n",
|
||||||
|
"import sys\n",
|
||||||
|
"\n",
|
||||||
|
"\n",
|
||||||
|
"N_TRN = 5000\n",
|
||||||
|
"N_TST = 5000\n",
|
||||||
|
"\n",
|
||||||
|
"DataBaseFolder = '/home/mchen/tmp/caffe/data/JStego/'\n",
|
||||||
|
"\n",
|
||||||
|
"BOSS2CoverPath = os.path.join( DataBaseFolder, '75') # BOSS2 cover subfolder\n",
|
||||||
|
"BOSS2StegoPath = os.path.join( DataBaseFolder, 'JUNI_0.4') # BOSS2 stego subfolder\n",
|
||||||
|
"\n",
|
||||||
|
"BOW2CoverPath = os.path.join( DataBaseFolder, 'BOWS2_75') # BOWS cover subfolder\n",
|
||||||
|
"BOW2StegoPath = os.path.join( DataBaseFolder, 'BOWS2_JUNI_0.4') # BOWS stego subfolder\n",
|
||||||
|
"\n",
|
||||||
|
"TxtListFolder = '/home/mchen/tmp/caffe/data/JStego/MiracleList/'\n",
|
||||||
|
"\n",
|
||||||
|
"np.random.seed(0) # reset the random seed\n",
|
||||||
|
"\n",
|
||||||
|
"RandomImages = np.random.permutation(10000) + 1\n",
|
||||||
|
"\n",
|
||||||
|
"print (\"[HG:] Train Set = %d, Test = %d\"%( 20000 + N_TRN * 2, N_TST * 2 ) )"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": 2,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true,
|
||||||
|
"deletable": true,
|
||||||
|
"editable": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"with open('{}/train.txt'.format(TxtListFolder), 'w') as f:\n",
|
||||||
|
" for idx in range(N_TRN):\n",
|
||||||
|
" imageName = str( RandomImages[idx] ) + '.jpg'\n",
|
||||||
|
" f.write('{} 0\\n'.format(BOSS2CoverPath + '/' + imageName ) )\n",
|
||||||
|
" f.write('{} 1\\n'.format(BOSS2StegoPath + '/' + imageName ) )\n",
|
||||||
|
" \n",
|
||||||
|
" for idx in range(10000):\n",
|
||||||
|
" imageName = str( RandomImages[idx] ) + '.jpg'\n",
|
||||||
|
" f.write('{} 0\\n'.format(BOW2CoverPath + '/' + imageName ) )\n",
|
||||||
|
" f.write('{} 1\\n'.format(BOW2StegoPath + '/' + imageName ) ) \n",
|
||||||
|
" \n",
|
||||||
|
" f.close()"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": 3,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true,
|
||||||
|
"deletable": true,
|
||||||
|
"editable": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"with open('{}/test.txt'.format(TxtListFolder), 'w') as f:\n",
|
||||||
|
" for idx in range(N_TST):\n",
|
||||||
|
" imageName = str( RandomImages[N_TRN + idx] ) + '.jpg'\n",
|
||||||
|
" f.write('{} 0\\n'.format(BOSS2CoverPath + '/' + imageName ) )\n",
|
||||||
|
" f.write('{} 1\\n'.format(BOSS2StegoPath + '/' + imageName ) )\n",
|
||||||
|
" f.close()"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true,
|
||||||
|
"deletable": true,
|
||||||
|
"editable": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": []
|
||||||
|
}
|
||||||
|
],
|
||||||
|
"metadata": {
|
||||||
|
"kernelspec": {
|
||||||
|
"display_name": "Python 2",
|
||||||
|
"language": "python",
|
||||||
|
"name": "python2"
|
||||||
|
},
|
||||||
|
"language_info": {
|
||||||
|
"codemirror_mode": {
|
||||||
|
"name": "ipython",
|
||||||
|
"version": 2
|
||||||
|
},
|
||||||
|
"file_extension": ".py",
|
||||||
|
"mimetype": "text/x-python",
|
||||||
|
"name": "python",
|
||||||
|
"nbconvert_exporter": "python",
|
||||||
|
"pygments_lexer": "ipython2",
|
||||||
|
"version": "2.7.6"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"nbformat": 4,
|
||||||
|
"nbformat_minor": 1
|
||||||
|
}
|
||||||
File diff suppressed because it is too large
Load Diff
|
|
@ -0,0 +1,17 @@
|
||||||
|
train_net: "/tmp/tmpNMRrso"
|
||||||
|
test_net: "/tmp/tmpwTrC5X"
|
||||||
|
test_iter: 1
|
||||||
|
test_interval: 1000000
|
||||||
|
base_lr: 0.0010000000475
|
||||||
|
display: 10
|
||||||
|
max_iter: 1000000
|
||||||
|
lr_policy: "step"
|
||||||
|
gamma: 0.75
|
||||||
|
momentum: 0.899999976158
|
||||||
|
weight_decay: 0.00999999977648
|
||||||
|
stepsize: 6000
|
||||||
|
snapshot: 6000
|
||||||
|
snapshot_prefix: "/home/mchen/tmp/caffe/examples/PhaseAwareNet"
|
||||||
|
solver_mode: GPU
|
||||||
|
iter_size: 1
|
||||||
|
type: "SGD"
|
||||||
|
|
@ -0,0 +1,463 @@
|
||||||
|
layer {
|
||||||
|
name: "data"
|
||||||
|
type: "Python"
|
||||||
|
top: "data"
|
||||||
|
top: "label"
|
||||||
|
python_param {
|
||||||
|
module: "AugStegoDataLayer"
|
||||||
|
layer: "AugmentDataLayerSync"
|
||||||
|
param_str: "{\'im_shape\': [512, 512], \'root\': \'/home/mchen/tmp/caffe/data/JStego/JUNI_0.4/\', \'split\': \'train\', \'batch_size\': 40}"
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "conv0"
|
||||||
|
type: "Convolution"
|
||||||
|
bottom: "data"
|
||||||
|
top: "conv0"
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
convolution_param {
|
||||||
|
num_output: 4
|
||||||
|
pad: 2
|
||||||
|
kernel_size: 5
|
||||||
|
stride: 1
|
||||||
|
weight_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "conv1"
|
||||||
|
type: "Convolution"
|
||||||
|
bottom: "conv0"
|
||||||
|
top: "conv1"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
convolution_param {
|
||||||
|
num_output: 8
|
||||||
|
pad: 2
|
||||||
|
kernel_size: 5
|
||||||
|
stride: 1
|
||||||
|
weight_filler {
|
||||||
|
type: "gaussian"
|
||||||
|
std: 0.00999999977648
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "abs1"
|
||||||
|
type: "AbsVal"
|
||||||
|
bottom: "conv1"
|
||||||
|
top: "abs1"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "bn1"
|
||||||
|
type: "BatchNorm"
|
||||||
|
bottom: "abs1"
|
||||||
|
top: "bn1"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
batch_norm_param {
|
||||||
|
moving_average_fraction: 0.980000019073
|
||||||
|
eps: 9.99999974738e-05
|
||||||
|
scale_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 1.0
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "tanh1"
|
||||||
|
type: "TanH"
|
||||||
|
bottom: "bn1"
|
||||||
|
top: "bn1"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "conv2"
|
||||||
|
type: "Convolution"
|
||||||
|
bottom: "bn1"
|
||||||
|
top: "conv2"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
convolution_param {
|
||||||
|
num_output: 16
|
||||||
|
pad: 2
|
||||||
|
kernel_size: 5
|
||||||
|
stride: 1
|
||||||
|
weight_filler {
|
||||||
|
type: "gaussian"
|
||||||
|
std: 0.00999999977648
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "bn2"
|
||||||
|
type: "BatchNorm"
|
||||||
|
bottom: "conv2"
|
||||||
|
top: "bn2"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
batch_norm_param {
|
||||||
|
moving_average_fraction: 0.980000019073
|
||||||
|
eps: 9.99999974738e-05
|
||||||
|
scale_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 1.0
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "tanh2"
|
||||||
|
type: "TanH"
|
||||||
|
bottom: "bn2"
|
||||||
|
top: "bn2"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "sbp"
|
||||||
|
type: "SplitByPhase"
|
||||||
|
bottom: "bn2"
|
||||||
|
top: "sbp"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "conv3"
|
||||||
|
type: "Convolution"
|
||||||
|
bottom: "sbp"
|
||||||
|
top: "conv3"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
convolution_param {
|
||||||
|
num_output: 128
|
||||||
|
pad: 0
|
||||||
|
kernel_size: 1
|
||||||
|
stride: 1
|
||||||
|
weight_filler {
|
||||||
|
type: "gaussian"
|
||||||
|
std: 0.00999999977648
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "bn3"
|
||||||
|
type: "BatchNorm"
|
||||||
|
bottom: "conv3"
|
||||||
|
top: "bn3"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
batch_norm_param {
|
||||||
|
moving_average_fraction: 0.980000019073
|
||||||
|
eps: 9.99999974738e-05
|
||||||
|
scale_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 1.0
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "relu3"
|
||||||
|
type: "ReLU"
|
||||||
|
bottom: "bn3"
|
||||||
|
top: "bn3"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "pool3"
|
||||||
|
type: "Pooling"
|
||||||
|
bottom: "bn3"
|
||||||
|
top: "pool3"
|
||||||
|
pooling_param {
|
||||||
|
pool: AVE
|
||||||
|
kernel_size: 5
|
||||||
|
stride: 2
|
||||||
|
pad: 1
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "conv4"
|
||||||
|
type: "Convolution"
|
||||||
|
bottom: "pool3"
|
||||||
|
top: "conv4"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
convolution_param {
|
||||||
|
num_output: 256
|
||||||
|
pad: 0
|
||||||
|
kernel_size: 1
|
||||||
|
stride: 1
|
||||||
|
weight_filler {
|
||||||
|
type: "gaussian"
|
||||||
|
std: 0.00999999977648
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "bn4"
|
||||||
|
type: "BatchNorm"
|
||||||
|
bottom: "conv4"
|
||||||
|
top: "bn4"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
batch_norm_param {
|
||||||
|
moving_average_fraction: 0.980000019073
|
||||||
|
eps: 9.99999974738e-05
|
||||||
|
scale_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 1.0
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "relu4"
|
||||||
|
type: "ReLU"
|
||||||
|
bottom: "bn4"
|
||||||
|
top: "bn4"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "pool4"
|
||||||
|
type: "Pooling"
|
||||||
|
bottom: "bn4"
|
||||||
|
top: "pool4"
|
||||||
|
pooling_param {
|
||||||
|
pool: AVE
|
||||||
|
kernel_size: 5
|
||||||
|
stride: 2
|
||||||
|
pad: 1
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "conv5"
|
||||||
|
type: "Convolution"
|
||||||
|
bottom: "pool4"
|
||||||
|
top: "conv5"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
convolution_param {
|
||||||
|
num_output: 512
|
||||||
|
pad: 0
|
||||||
|
kernel_size: 1
|
||||||
|
stride: 1
|
||||||
|
weight_filler {
|
||||||
|
type: "gaussian"
|
||||||
|
std: 0.00999999977648
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "bn5"
|
||||||
|
type: "BatchNorm"
|
||||||
|
bottom: "conv5"
|
||||||
|
top: "bn5"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 0.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
batch_norm_param {
|
||||||
|
moving_average_fraction: 0.980000019073
|
||||||
|
eps: 9.99999974738e-05
|
||||||
|
scale_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 1.0
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.0
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "relu5"
|
||||||
|
type: "ReLU"
|
||||||
|
bottom: "bn5"
|
||||||
|
top: "bn5"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "pool5"
|
||||||
|
type: "Pooling"
|
||||||
|
bottom: "bn5"
|
||||||
|
top: "pool5"
|
||||||
|
pooling_param {
|
||||||
|
pool: AVE
|
||||||
|
global_pooling: true
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "fc6"
|
||||||
|
type: "InnerProduct"
|
||||||
|
bottom: "pool5"
|
||||||
|
top: "fc6"
|
||||||
|
param {
|
||||||
|
lr_mult: 1.0
|
||||||
|
decay_mult: 1.0
|
||||||
|
}
|
||||||
|
param {
|
||||||
|
lr_mult: 2.0
|
||||||
|
decay_mult: 0.0
|
||||||
|
}
|
||||||
|
inner_product_param {
|
||||||
|
num_output: 2
|
||||||
|
weight_filler {
|
||||||
|
type: "xavier"
|
||||||
|
}
|
||||||
|
bias_filler {
|
||||||
|
type: "constant"
|
||||||
|
value: 0.00999999977648
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "loss"
|
||||||
|
type: "SoftmaxWithLoss"
|
||||||
|
bottom: "fc6"
|
||||||
|
bottom: "label"
|
||||||
|
top: "loss"
|
||||||
|
}
|
||||||
|
layer {
|
||||||
|
name: "acc"
|
||||||
|
type: "Accuracy"
|
||||||
|
bottom: "fc6"
|
||||||
|
bottom: "label"
|
||||||
|
top: "acc"
|
||||||
|
}
|
||||||
Binary file not shown.
|
|
@ -0,0 +1,76 @@
|
||||||
|
#include <algorithm>
|
||||||
|
#include <vector>
|
||||||
|
|
||||||
|
#include "caffe/filler.hpp"
|
||||||
|
#include "caffe/layer_factory.hpp"
|
||||||
|
#include "caffe/layers/split_by_phase_layer.hpp"
|
||||||
|
#include "caffe/util/math_functions.hpp"
|
||||||
|
|
||||||
|
namespace caffe {
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
void SplitByPhaseLayer<Dtype>::LayerSetUp(const vector<Blob<Dtype>*>& bottom,
|
||||||
|
const vector<Blob<Dtype>*>& top) {
|
||||||
|
|
||||||
|
num_images_ = bottom[0]->num();
|
||||||
|
num_filters_ = bottom[0]->channels();
|
||||||
|
height_ = bottom[0]->height();
|
||||||
|
width_ = bottom[0]->width();
|
||||||
|
|
||||||
|
CHECK_EQ(height_, 512);
|
||||||
|
CHECK_EQ(width_, 512);
|
||||||
|
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
void SplitByPhaseLayer<Dtype>::Reshape(const vector<Blob<Dtype>*>& bottom,
|
||||||
|
const vector<Blob<Dtype>*>& top) {
|
||||||
|
|
||||||
|
top[0]->Reshape(num_images_, num_filters_*64, 64, 64);
|
||||||
|
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
void SplitByPhaseLayer<Dtype>::Forward_cpu(
|
||||||
|
const vector<Blob<Dtype>*>& bottom, const vector<Blob<Dtype>*>& top) {
|
||||||
|
const Dtype* bottom_data = bottom[0]->cpu_data();
|
||||||
|
Dtype* top_data = top[0]->mutable_cpu_data();
|
||||||
|
int n, c, p, h, w, source_index;
|
||||||
|
for (int index = 0; index < bottom[0]->count(); ++index) {
|
||||||
|
w = index % 64;
|
||||||
|
h = (index / 64) % 64;
|
||||||
|
p = (index / 64 / 64) % 64;
|
||||||
|
c = (index / 64 / 64 / 64) % num_filters_;
|
||||||
|
n = index / 64 / 64 / 64 / num_filters_;
|
||||||
|
source_index = ((w*8)+(h*8*512)+(p%8)+(p/8)*512)+((n*num_filters_+c)*512*512);
|
||||||
|
top_data[index] = bottom_data[source_index];
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
void SplitByPhaseLayer<Dtype>::Backward_cpu(const vector<Blob<Dtype>*>& top,
|
||||||
|
const vector<bool>& propagate_down, const vector<Blob<Dtype>*>& bottom) {
|
||||||
|
if (propagate_down[0]) {
|
||||||
|
const Dtype* top_diff = top[0]->cpu_diff();
|
||||||
|
Dtype* bottom_diff = bottom[0]->mutable_cpu_diff();
|
||||||
|
int n, c, p, h, w, source_index;
|
||||||
|
for (int index = 0; index < bottom[0]->count(); ++index) {
|
||||||
|
w = index % 64;
|
||||||
|
h = (index / 64) % 64;
|
||||||
|
p = (index / 64 / 64) % 64;
|
||||||
|
c = (index / 64 / 64 / 64) % num_filters_;
|
||||||
|
n = index / 64 / 64 / 64 / num_filters_;
|
||||||
|
source_index = ((w*8)+(h*8*512)+(p%8)+(p/8)*512)+((n*num_filters_+c)*512*512);
|
||||||
|
bottom_diff[source_index] = top_diff[index];
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
#ifdef CPU_ONLY
|
||||||
|
STUB_GPU(SplitByPhaseLayer);
|
||||||
|
#endif
|
||||||
|
|
||||||
|
INSTANTIATE_CLASS(SplitByPhaseLayer);
|
||||||
|
REGISTER_LAYER_CLASS(SplitByPhase);
|
||||||
|
|
||||||
|
} // namespace caffe
|
||||||
|
|
@ -0,0 +1,92 @@
|
||||||
|
#include <cfloat>
|
||||||
|
#include <vector>
|
||||||
|
|
||||||
|
#include "caffe/layers/split_by_phase_layer.hpp"
|
||||||
|
#include "caffe/util/math_functions.hpp"
|
||||||
|
|
||||||
|
namespace caffe {
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
__global__ void SplitByPhaseForward(const int nthreads,
|
||||||
|
const Dtype* const bottom_data, const int num_filters, Dtype* const top_data) {
|
||||||
|
CUDA_KERNEL_LOOP(index, nthreads) {
|
||||||
|
const int w = index % 64;
|
||||||
|
const int h = (index / 64) % 64;
|
||||||
|
const int p = (index / 64 / 64) % 64;
|
||||||
|
const int c = (index / 64 / 64 / 64) % num_filters;
|
||||||
|
const int n = index / 64 / 64 / 64 / num_filters;
|
||||||
|
const int source_index = ((w*8)+(h*8*512)+(p%8)+(p/8)*512)+((n*num_filters+c)*512*512);
|
||||||
|
top_data[index] = bottom_data[source_index];;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
__global__ void SplitByPhaseForwardSlow(const int nthreads,
|
||||||
|
const Dtype* const bottom_data, const int num_filters, Dtype* const top_data) {
|
||||||
|
CUDA_KERNEL_LOOP(index, nthreads) {
|
||||||
|
int h, p, c, n, source_index;
|
||||||
|
for (int w = 0; w < 64; ++w) {
|
||||||
|
h = index % 64;
|
||||||
|
p = (index / 64) % 64;
|
||||||
|
c = (index / 64 / 64) % num_filters;
|
||||||
|
n = index / 64 / 64 / num_filters;
|
||||||
|
source_index = ((w*8)+(h*8*512)+(p%8)+(p/8)*512)+((n*num_filters+c)*512*512);
|
||||||
|
top_data[index*64+w] = bottom_data[source_index];
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
void SplitByPhaseLayer<Dtype>::Forward_gpu(
|
||||||
|
const vector<Blob<Dtype>*>& bottom, const vector<Blob<Dtype>*>& top) {
|
||||||
|
const Dtype* bottom_data = bottom[0]->gpu_data();
|
||||||
|
Dtype* top_data = top[0]->mutable_gpu_data();
|
||||||
|
const int count = top[0]->count();
|
||||||
|
// NOLINT_NEXT_LINE(whitespace/operators)
|
||||||
|
SplitByPhaseForward<Dtype><<<CAFFE_GET_BLOCKS(count), CAFFE_CUDA_NUM_THREADS>>>(
|
||||||
|
count, bottom_data, num_filters_, top_data);
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
__global__ void SplitByPhaseBackwardSlow(const int nthreads,
|
||||||
|
Dtype* const bottom_diff, const int num_filters, const Dtype* const top_diff) {
|
||||||
|
CUDA_KERNEL_LOOP(index, nthreads) {
|
||||||
|
const int w = index % 64;
|
||||||
|
const int h = (index / 64) % 64;
|
||||||
|
const int p = (index / 64 / 64) % 64;
|
||||||
|
const int c = (index / 64 / 64 / 64) % num_filters;
|
||||||
|
const int n = index / 64 / 64 / 64 / num_filters;
|
||||||
|
const int source_index = ((w*8)+(h*8*512)+(p%8)+(p/8)*512)+((n*num_filters+c)*512*512);
|
||||||
|
bottom_diff[source_index] = top_diff[index];
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
__global__ void SplitByPhaseBackward(const int nthreads,
|
||||||
|
Dtype* const bottom_diff, const int num_filters, const Dtype* const top_diff) {
|
||||||
|
CUDA_KERNEL_LOOP(index, nthreads) {
|
||||||
|
const int w = index % 512;
|
||||||
|
const int h = (index / 512) % 512;
|
||||||
|
const int c = (index / 512 / 512) % num_filters;
|
||||||
|
const int n = index / 512 / 512 / num_filters;
|
||||||
|
const int target_index = ((w/8)+64*(h/8))+(64*64*(((w%8)+8*(h%8))))+(512*512*(n*num_filters+c));
|
||||||
|
bottom_diff[index] = top_diff[target_index];
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
void SplitByPhaseLayer<Dtype>::Backward_gpu(const vector<Blob<Dtype>*>& top,
|
||||||
|
const vector<bool>& propagate_down, const vector<Blob<Dtype>*>& bottom) {
|
||||||
|
if (propagate_down[0]) {
|
||||||
|
const Dtype* top_diff = top[0]->gpu_diff();
|
||||||
|
Dtype* bottom_diff = bottom[0]->mutable_gpu_diff();
|
||||||
|
const int count = bottom[0]->count();
|
||||||
|
// NOLINT_NEXT_LINE(whitespace/operators)
|
||||||
|
SplitByPhaseBackward<Dtype><<<CAFFE_GET_BLOCKS(count), CAFFE_CUDA_NUM_THREADS>>>(
|
||||||
|
count, bottom_diff, num_filters_, top_diff);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
INSTANTIATE_LAYER_GPU_FUNCS(SplitByPhaseLayer);
|
||||||
|
|
||||||
|
} // namespace caffe
|
||||||
|
|
@ -0,0 +1,57 @@
|
||||||
|
#ifndef CAFFE_SPLIT_BY_PHASE_LAYER_HPP_
|
||||||
|
#define CAFFE_SPLIT_BY_PHASE_LAYER_HPP_
|
||||||
|
|
||||||
|
#include <vector>
|
||||||
|
|
||||||
|
#include "caffe/blob.hpp"
|
||||||
|
#include "caffe/layer.hpp"
|
||||||
|
#include "caffe/proto/caffe.pb.h"
|
||||||
|
|
||||||
|
namespace caffe {
|
||||||
|
|
||||||
|
template <typename Dtype>
|
||||||
|
class SplitByPhaseLayer: public Layer<Dtype> {
|
||||||
|
public:
|
||||||
|
explicit SplitByPhaseLayer(const LayerParameter& param)
|
||||||
|
: Layer<Dtype>(param) {}
|
||||||
|
virtual void LayerSetUp(const vector<Blob<Dtype>*>& bottom,
|
||||||
|
const vector<Blob<Dtype>*>& top);
|
||||||
|
virtual void Reshape(const vector<Blob<Dtype>*>& bottom,
|
||||||
|
const vector<Blob<Dtype>*>& top);
|
||||||
|
|
||||||
|
virtual inline const char* type() const { return "SplitByPhase"; }
|
||||||
|
virtual inline int ExactNumBottomBlobs() const { return 1; }
|
||||||
|
virtual inline int ExactNumTopBlobs() const { return 1; }
|
||||||
|
|
||||||
|
protected:
|
||||||
|
virtual void Forward_cpu(const vector<Blob<Dtype>*>& bottom,
|
||||||
|
const vector<Blob<Dtype>*>& top);
|
||||||
|
virtual void Forward_gpu(const vector<Blob<Dtype>*>& bottom,
|
||||||
|
const vector<Blob<Dtype>*>& top);
|
||||||
|
virtual void Backward_cpu(const vector<Blob<Dtype>*>& top,
|
||||||
|
const vector<bool>& propagate_down, const vector<Blob<Dtype>*>& bottom);
|
||||||
|
virtual void Backward_gpu(const vector<Blob<Dtype>*>& top,
|
||||||
|
const vector<bool>& propagate_down, const vector<Blob<Dtype>*>& bottom);
|
||||||
|
|
||||||
|
shared_ptr<Layer<Dtype> > bias_layer_;
|
||||||
|
vector<Blob<Dtype>*> bias_bottom_vec_;
|
||||||
|
vector<bool> bias_propagate_down_;
|
||||||
|
int bias_param_id_;
|
||||||
|
|
||||||
|
Blob<Dtype> sum_multiplier_;
|
||||||
|
Blob<Dtype> sum_result_;
|
||||||
|
Blob<Dtype> temp_;
|
||||||
|
int axis_;
|
||||||
|
int outer_dim_, scale_dim_, inner_dim_;
|
||||||
|
|
||||||
|
int num_images_;
|
||||||
|
int num_filters_;
|
||||||
|
int height_;
|
||||||
|
int width_;
|
||||||
|
|
||||||
|
};
|
||||||
|
|
||||||
|
|
||||||
|
} // namespace caffe
|
||||||
|
|
||||||
|
#endif // CAFFE_SPLIT_BY_PHASE_LAYER_HPP_
|
||||||
|
|
@ -0,0 +1,123 @@
|
||||||
|
#include <vector>
|
||||||
|
|
||||||
|
#include "gtest/gtest.h"
|
||||||
|
|
||||||
|
#include "caffe/blob.hpp"
|
||||||
|
#include "caffe/common.hpp"
|
||||||
|
#include "caffe/filler.hpp"
|
||||||
|
#include "caffe/layers/split_by_phase_layer.hpp"
|
||||||
|
|
||||||
|
#include "caffe/test/test_caffe_main.hpp"
|
||||||
|
#include "caffe/test/test_gradient_check_util.hpp"
|
||||||
|
|
||||||
|
namespace caffe {
|
||||||
|
|
||||||
|
#ifndef CPU_ONLY
|
||||||
|
extern cudaDeviceProp CAFFE_TEST_CUDA_PROP;
|
||||||
|
#endif
|
||||||
|
|
||||||
|
template <typename TypeParam>
|
||||||
|
class SplitByPhaseLayerTest : public MultiDeviceTest<TypeParam> {
|
||||||
|
typedef typename TypeParam::Dtype Dtype;
|
||||||
|
protected:
|
||||||
|
SplitByPhaseLayerTest()
|
||||||
|
: blob_bottom_(new Blob<Dtype>(1, 1, 512, 512)),
|
||||||
|
blob_bottom_nobatch_(new Blob<Dtype>(1, 2, 3, 4)),
|
||||||
|
blob_top_(new Blob<Dtype>()) {
|
||||||
|
// fill the values
|
||||||
|
FillerParameter filler_param;
|
||||||
|
// filler_param.s
|
||||||
|
filler_param.set_min(-9);
|
||||||
|
filler_param.set_max(9);
|
||||||
|
UniformFiller<Dtype> filler(filler_param);
|
||||||
|
filler.Fill(this->blob_bottom_);
|
||||||
|
blob_top_vec_.push_back(blob_top_);
|
||||||
|
}
|
||||||
|
virtual ~SplitByPhaseLayerTest() {
|
||||||
|
delete blob_bottom_;
|
||||||
|
delete blob_bottom_nobatch_;
|
||||||
|
delete blob_top_;
|
||||||
|
}
|
||||||
|
Blob<Dtype>* const blob_bottom_;
|
||||||
|
Blob<Dtype>* const blob_bottom_nobatch_;
|
||||||
|
Blob<Dtype>* const blob_top_;
|
||||||
|
vector<Blob<Dtype>*> blob_bottom_vec_;
|
||||||
|
vector<Blob<Dtype>*> blob_top_vec_;
|
||||||
|
};
|
||||||
|
|
||||||
|
TYPED_TEST_CASE(SplitByPhaseLayerTest, TestDtypesAndDevices);
|
||||||
|
|
||||||
|
TYPED_TEST(SplitByPhaseLayerTest, TestSetUp) {
|
||||||
|
typedef typename TypeParam::Dtype Dtype;
|
||||||
|
this->blob_bottom_vec_.push_back(this->blob_bottom_);
|
||||||
|
LayerParameter layer_param;
|
||||||
|
shared_ptr<SplitByPhaseLayer<Dtype> > layer(
|
||||||
|
new SplitByPhaseLayer<Dtype>(layer_param));
|
||||||
|
layer->SetUp(this->blob_bottom_vec_, this->blob_top_vec_);
|
||||||
|
EXPECT_EQ(this->blob_top_vec_[0]->num(), 1);
|
||||||
|
EXPECT_EQ(this->blob_top_vec_[0]->channels(), 1*64);
|
||||||
|
EXPECT_EQ(this->blob_top_vec_[0]->height(), 64);
|
||||||
|
EXPECT_EQ(this->blob_top_vec_[0]->width(), 64);
|
||||||
|
}
|
||||||
|
|
||||||
|
TYPED_TEST(SplitByPhaseLayerTest, TestForward) {
|
||||||
|
typedef typename TypeParam::Dtype Dtype;
|
||||||
|
Dtype* bottom_data = this->blob_bottom_->mutable_cpu_data();
|
||||||
|
const int num_filters = this->blob_bottom_->channels();
|
||||||
|
int n, c, p, h, w, bottom_fill_idx;
|
||||||
|
for (int index = 0; index < this->blob_bottom_->count(); ++index) {
|
||||||
|
w = index % 64;
|
||||||
|
h = (index / 64) % 64;
|
||||||
|
p = (index / 64 / 64) % 64;
|
||||||
|
c = (index / 64 / 64 / 64) % num_filters;
|
||||||
|
n = index / 64 / 64 / 64 / num_filters;
|
||||||
|
bottom_fill_idx = ((w*8)+(h*8*512)+(p%8)+(p/8)*512)+((n*num_filters+c)*512*512);
|
||||||
|
bottom_data[bottom_fill_idx] = p;
|
||||||
|
}
|
||||||
|
this->blob_bottom_vec_.push_back(this->blob_bottom_);
|
||||||
|
bool IS_VALID_CUDA = false;
|
||||||
|
#ifndef CPU_ONLY
|
||||||
|
IS_VALID_CUDA = CAFFE_TEST_CUDA_PROP.major >= 2;
|
||||||
|
#endif
|
||||||
|
if (Caffe::mode() == Caffe::CPU ||
|
||||||
|
sizeof(Dtype) == 4 || IS_VALID_CUDA) {
|
||||||
|
LayerParameter layer_param;
|
||||||
|
shared_ptr<SplitByPhaseLayer<Dtype> > layer(
|
||||||
|
new SplitByPhaseLayer<Dtype>(layer_param));
|
||||||
|
layer->SetUp(this->blob_bottom_vec_, this->blob_top_vec_);
|
||||||
|
layer->Forward(this->blob_bottom_vec_, this->blob_top_vec_);
|
||||||
|
const Dtype* data = this->blob_top_vec_[0]->cpu_data();
|
||||||
|
const int num_phase_blocks = this->blob_top_vec_[0]->num()*this->blob_top_vec_[0]->channels();
|
||||||
|
for (int nc = 0; nc < num_phase_blocks; ++nc) {
|
||||||
|
for (int h = 0; h < 64 ; ++h ) {
|
||||||
|
for (int w = 0; w < 64 ; ++w ) {
|
||||||
|
CHECK_EQ(data[nc*(64*64)+h*64+w], nc%64);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
} else {
|
||||||
|
LOG(ERROR) << "Skipping test due to old architecture.";
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
TYPED_TEST(SplitByPhaseLayerTest, TestGradient) {
|
||||||
|
typedef typename TypeParam::Dtype Dtype;
|
||||||
|
this->blob_bottom_vec_.push_back(this->blob_bottom_);
|
||||||
|
bool IS_VALID_CUDA = false;
|
||||||
|
#ifndef CPU_ONLY
|
||||||
|
IS_VALID_CUDA = CAFFE_TEST_CUDA_PROP.major >= 2;
|
||||||
|
#endif
|
||||||
|
// if (Caffe::mode() == Caffe::CPU ||
|
||||||
|
// sizeof(Dtype) == 4 || IS_VALID_CUDA) {
|
||||||
|
if (Caffe::mode() == Caffe::GPU) {
|
||||||
|
LayerParameter layer_param;
|
||||||
|
SplitByPhaseLayer<Dtype> layer(layer_param);
|
||||||
|
GradientChecker<Dtype> checker(1e-2, 1e-3);
|
||||||
|
checker.CheckGradientExhaustive(&layer, this->blob_bottom_vec_,
|
||||||
|
this->blob_top_vec_);
|
||||||
|
} else {
|
||||||
|
LOG(ERROR) << "Skipping test due to old architecture.";
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
} // namespace caffe
|
||||||
Binary file not shown.
|
After Width: | Height: | Size: 191 KiB |
Binary file not shown.
|
After Width: | Height: | Size: 191 KiB |
|
|
@ -0,0 +1,70 @@
|
||||||
|
function [BN_Moments, stats] = bn_refine_phaseaware(varargin)
|
||||||
|
% Test the phasesplit net
|
||||||
|
|
||||||
|
opts.batchSize = 48;
|
||||||
|
opts.expDir = fullfile('data', 'JUNI-7504-PNet-dagnn-40-Seed-0-log_short') ;
|
||||||
|
opts.testEpoch = 40;
|
||||||
|
opts.saveResult = true;
|
||||||
|
opts.bnEpochCollectSize = 2000;
|
||||||
|
opts.gpuIdx = 1;
|
||||||
|
|
||||||
|
opts = vl_argparse( opts, varargin );
|
||||||
|
|
||||||
|
opts.imdbPath = fullfile(opts.expDir, 'imdb.mat');
|
||||||
|
|
||||||
|
opts.train = struct('gpus', opts.gpuIdx, 'cudnn', true, 'stegoShuffle', true ) ;
|
||||||
|
if ~isfield(opts.train, 'gpus'), opts.train.gpus = []; end;
|
||||||
|
|
||||||
|
% put it to drawing
|
||||||
|
if ( ~exist( opts.expDir, 'dir' ) )
|
||||||
|
error('expDir is empty' );
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Find the data base
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
if exist(opts.imdbPath, 'file')
|
||||||
|
imdb = load(opts.imdbPath) ;
|
||||||
|
else
|
||||||
|
error(' cannot find imdb' );
|
||||||
|
end
|
||||||
|
|
||||||
|
|
||||||
|
meta.inputSize = [512, 512, 1, opts.batchSize];
|
||||||
|
|
||||||
|
[BN_Moments, stats] = cnn_bnrefine_dag(imdb, getBatchFn( opts, meta ), ...
|
||||||
|
'expDir', opts.expDir, ...
|
||||||
|
'batchSize', opts.batchSize, ...
|
||||||
|
'testEpoch', opts.testEpoch, ...
|
||||||
|
'bnEpochCollectSize', opts.bnEpochCollectSize, ...
|
||||||
|
'saveResult', opts.saveResult, ...
|
||||||
|
opts.train ) ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function fn = getBatchFn(opts, meta)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
bopts.useGpu = numel(opts.train.gpus) > 0 ;
|
||||||
|
bopts.imageSize = meta.inputSize;
|
||||||
|
|
||||||
|
fn = @(x,y) getDagNNBatch(bopts,x,y) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function inputs = getDagNNBatch(opts, imdb, batch)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% label
|
||||||
|
labels = imdb.images.label(1,batch) ;
|
||||||
|
% images
|
||||||
|
images = zeros(opts.imageSize(1), opts.imageSize(2), ...
|
||||||
|
opts.imageSize(3), numel(batch), 'single') ;
|
||||||
|
for i = 1:numel(batch)
|
||||||
|
imt = load(imdb.images.name{batch(i)}, 'im');
|
||||||
|
images(:,:,:,i) = single(imt.im);
|
||||||
|
% imt = imread(imdb.images.name{batch(i)});
|
||||||
|
% images(:,:,:,i) = single(imt);
|
||||||
|
end
|
||||||
|
|
||||||
|
if opts.useGpu > 0
|
||||||
|
images = gpuArray(images) ;
|
||||||
|
end
|
||||||
|
inputs = {'input', images, 'label', labels} ;
|
||||||
|
|
@ -0,0 +1,149 @@
|
||||||
|
function [net, info] = cnn_phaseaware(varargin)
|
||||||
|
%CNN_PHASEAWARE Demonstrates training a PhaseAwareNet on JUNI and UED
|
||||||
|
|
||||||
|
run(fullfile(fileparts(mfilename('fullpath')), ...
|
||||||
|
'..', '..', 'matlab', 'vl_setupnn.m')) ;
|
||||||
|
|
||||||
|
opts.modelType = 'PNet';
|
||||||
|
opts.seed = 0;
|
||||||
|
opts.networkType = 'dagnn' ;
|
||||||
|
opts.batchSize = 40;
|
||||||
|
opts.lrSequence = 'log_short';
|
||||||
|
opts.printDotFile = true;
|
||||||
|
opts.coverPath = 'C:\DeepLearning\matconvnet-1.0-beta20\data\JStego\75_mat';
|
||||||
|
opts.stegoPath = 'C:\DeepLearning\matconvnet-1.0-beta20\data\JStego\JUNI_0.4_mat';
|
||||||
|
|
||||||
|
|
||||||
|
sfx = [opts.modelType, '-', opts.networkType, '-', num2str(opts.batchSize), ...
|
||||||
|
'-Seed-', num2str(opts.seed), '-', opts.lrSequence] ;
|
||||||
|
opts.expDir = fullfile('data', ['JUNI-7504-' sfx]) ; % TODO
|
||||||
|
|
||||||
|
opts.imdbPath = fullfile(opts.expDir, 'imdb.mat');
|
||||||
|
|
||||||
|
opts.train = struct('gpus', [1,2], 'cudnn', true, 'stegoShuffle', true, 'computeBNMoment', true) ;
|
||||||
|
if ~isfield(opts.train, 'gpus'), opts.train.gpus = []; end;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Prepare model
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
if (strcmpi( opts.modelType, 'PNet' ))
|
||||||
|
|
||||||
|
net = cnn_phaseaware_PNet_init( 'networkType', opts.networkType, ...
|
||||||
|
'batchSize', opts.batchSize, ...
|
||||||
|
'seed', opts.seed, ...
|
||||||
|
'lrSequence', opts.lrSequence );
|
||||||
|
|
||||||
|
elseif (strcmpi( opts.modelType, 'VNet' ))
|
||||||
|
|
||||||
|
net = cnn_phaseaware_VNet_init( 'networkType', opts.networkType, ...
|
||||||
|
'batchSize', opts.batchSize, ...
|
||||||
|
'seed', opts.seed, ...
|
||||||
|
'lrSequence', opts.lrSequence );
|
||||||
|
|
||||||
|
else
|
||||||
|
error('Unknown model type');
|
||||||
|
end
|
||||||
|
|
||||||
|
% put it to drawing
|
||||||
|
if ( ~exist( opts.expDir, 'dir' ) )
|
||||||
|
mkdir( opts.expDir );
|
||||||
|
end
|
||||||
|
|
||||||
|
if opts.printDotFile
|
||||||
|
net2dot(net, fullfile( opts.expDir, 'NetConfig.dot' ), ...
|
||||||
|
'BatchSize', net.meta.trainOpts.batchSize, ...
|
||||||
|
'Inputs', {'input', [net.meta.inputSize, net.meta.trainOpts.batchSize]});
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Prepare data
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
if exist(opts.imdbPath, 'file')
|
||||||
|
imdb = load(opts.imdbPath) ;
|
||||||
|
else
|
||||||
|
imdb = cnn_phaseaware_imdb_setup('coverPath', opts.coverPath, 'stegoPath', opts.stegoPath) ;
|
||||||
|
|
||||||
|
save(opts.imdbPath, '-struct', 'imdb') ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% Set the class names in the network
|
||||||
|
net.meta.classes.name = imdb.classes.name ;
|
||||||
|
net.meta.classes.description = imdb.classes.description ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Learn
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
switch opts.networkType
|
||||||
|
case 'dagnn', trainFn = @cnn_train_dag ;
|
||||||
|
otherwise, error('wrong network type');
|
||||||
|
end
|
||||||
|
|
||||||
|
[net, info] = trainFn(net, imdb, getBatchFn(opts, net.meta), ...
|
||||||
|
'expDir', opts.expDir, ...
|
||||||
|
net.meta.trainOpts, ...
|
||||||
|
opts.train) ;
|
||||||
|
|
||||||
|
modelPath = fullfile(opts.expDir, 'net-deployed.mat');
|
||||||
|
|
||||||
|
switch opts.networkType
|
||||||
|
case 'dagnn'
|
||||||
|
net_ = net.saveobj() ;
|
||||||
|
save(modelPath, '-struct', 'net_') ;
|
||||||
|
clear net_ ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function fn = getBatchFn(opts, meta)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
bopts.useGpu = numel(opts.train.gpus) > 0 ;
|
||||||
|
bopts.imageSize = meta.inputSize;
|
||||||
|
|
||||||
|
switch lower(opts.networkType)
|
||||||
|
case 'dagnn'
|
||||||
|
fn = @(x,y) getDagNNBatch(bopts,x,y) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function inputs = getDagNNBatch(opts, imdb, batch)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% label
|
||||||
|
labels = imdb.images.label(1,batch) ;
|
||||||
|
% images
|
||||||
|
images = zeros(opts.imageSize(1), opts.imageSize(2), ...
|
||||||
|
opts.imageSize(3), numel(batch), 'single') ;
|
||||||
|
|
||||||
|
for i = 1:numel(batch)/2
|
||||||
|
|
||||||
|
% cover = imread(imdb.images.name{batch(2*i-1)});
|
||||||
|
% stego = imread(imdb.images.name{batch(2*i)});
|
||||||
|
|
||||||
|
imt = load(imdb.images.name{batch(2*i-1)}, 'im');
|
||||||
|
cover = single(imt.im);
|
||||||
|
|
||||||
|
imt = load(imdb.images.name{batch(2*i)}, 'im');
|
||||||
|
stego = single(imt.im);
|
||||||
|
|
||||||
|
% random rotate, 0, 90, 180, 270
|
||||||
|
r = randi(4) - 1;
|
||||||
|
cover = rot90( cover, r );
|
||||||
|
stego = rot90( stego, r );
|
||||||
|
|
||||||
|
% random mirror flip
|
||||||
|
if ( rand > 0.5 )
|
||||||
|
cover = fliplr( cover );
|
||||||
|
stego = fliplr( stego );
|
||||||
|
end
|
||||||
|
|
||||||
|
images(:,:,:,2*i-1) = single(cover);
|
||||||
|
images(:,:,:,2*i) = single(stego);
|
||||||
|
|
||||||
|
end
|
||||||
|
|
||||||
|
if opts.useGpu > 0
|
||||||
|
images = gpuArray(images) ;
|
||||||
|
end
|
||||||
|
inputs = {'input', images, 'label', labels} ;
|
||||||
|
|
||||||
|
|
||||||
|
|
@ -0,0 +1,265 @@
|
||||||
|
function net = cnn_phaseaware_PNet_init(varargin)
|
||||||
|
% Define and initialize PhaseAwareNet net
|
||||||
|
opts.networkType = 'dagnn' ;
|
||||||
|
opts.batchSize = 40;
|
||||||
|
opts.seed = 0;
|
||||||
|
opts.lrSequence = 'step_long2';
|
||||||
|
opts = vl_argparse(opts, varargin) ;
|
||||||
|
|
||||||
|
rng( opts.seed );
|
||||||
|
|
||||||
|
net.layers = {} ;
|
||||||
|
|
||||||
|
convOpts = {'CudnnWorkspaceLimit', 1024*1024*1204} ;
|
||||||
|
|
||||||
|
HPF = zeros(5, 5, 1, 4, 'single');
|
||||||
|
|
||||||
|
HPF(:,:,1,1) = [ -1, 2, -2, 2, -1; ...
|
||||||
|
2, -6, 8, -6, 2; ...
|
||||||
|
-2, 8, -12, 8, -2; ...
|
||||||
|
2, -6, 8, -6, 2; ...
|
||||||
|
-1, 2, -2, 2, -1]/12;
|
||||||
|
|
||||||
|
HPF(:,:,1,2) = [ 0, 0, 5.2, 0, 0; ...
|
||||||
|
0, 23.4, 36.4, 23.4, 0; ...
|
||||||
|
5.2, 36.4, -261.0, 36.4, 5.2; ...
|
||||||
|
0, 23.4, 36.4, 23.4, 0; ...
|
||||||
|
0, 0, 5.2, 0, 0]/261;
|
||||||
|
|
||||||
|
HPF(:,:,1,3) = [ 0.0562, -0.1354, 0.0000, 0.1354, -0.0562; ...
|
||||||
|
0.0818, -0.1970, 0.0000, 0.1970, -0.0818; ...
|
||||||
|
0.0926, -0.2233, 0.0000, 0.2233, -0.0926; ...
|
||||||
|
0.0818, -0.1970, 0.0000, 0.1970, -0.0818; ...
|
||||||
|
0.0562, -0.1354, 0.0000, 0.1354, -0.0562 ];
|
||||||
|
|
||||||
|
HPF(:,:,1,4) = [-0.0562, -0.0818, -0.0926, -0.0818, -0.0562; ...
|
||||||
|
0.1354, 0.1970, 0.2233, 0.1970, 0.1354; ...
|
||||||
|
0.0000, 0.0000, 0.0000, -0.0000, -0.0000; ...
|
||||||
|
-0.1354, -0.1970, -0.2233, -0.1970, -0.1354; ...
|
||||||
|
0.0562, 0.0818, 0.0926, 0.0818, 0.0562 ];
|
||||||
|
|
||||||
|
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'HPFs', ...
|
||||||
|
'weights', {{HPF, []}}, ...
|
||||||
|
'learningRate', [0, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 2, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
|
||||||
|
% Group 1
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_1', ...
|
||||||
|
'weights', {{init_weight('gaussian', 5, 5, 4, 8, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 2, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'abs', 'name', 'ABS_1') ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_1', ...
|
||||||
|
'weights', {{ones(8, 1, 'single'), ...
|
||||||
|
zeros(8, 1, 'single'), ...
|
||||||
|
zeros(8, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'tanh', 'name', 'TanH_1') ;
|
||||||
|
|
||||||
|
|
||||||
|
% Group 2
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_2', ...
|
||||||
|
'weights', {{init_weight('gaussian', 5, 5, 8, 16, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 2, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_2', ...
|
||||||
|
'weights', {{ones(16, 1, 'single'), ...
|
||||||
|
zeros(16, 1, 'single'), ...
|
||||||
|
zeros(16, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'tanh', 'name', 'TanH_2') ;
|
||||||
|
|
||||||
|
|
||||||
|
% Phase split here
|
||||||
|
net.layers{end+1} = struct('type', 'phasesplit', ...
|
||||||
|
'name', 'DCTPhaseSplit', ...
|
||||||
|
'pool', [1, 1], ...
|
||||||
|
'stride', 8, ...
|
||||||
|
'pad', 0 );
|
||||||
|
|
||||||
|
DCTMode = 64;
|
||||||
|
% Group 3
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_3', ...
|
||||||
|
'weights', {{init_weight('gaussian', 1, 1, 16, 32*DCTMode, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_3', ...
|
||||||
|
'weights', {{ones(32*DCTMode, 1, 'single'), ...
|
||||||
|
zeros(32*DCTMode, 1, 'single'), ...
|
||||||
|
zeros(32*DCTMode, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'relu', 'name', 'ReLU_3') ;
|
||||||
|
net.layers{end+1} = struct('type', 'pool', ...
|
||||||
|
'name', 'Pool_3', ...
|
||||||
|
'method', 'avg', ...
|
||||||
|
'pool', [5 5], ...
|
||||||
|
'stride', 2, ...
|
||||||
|
'pad', 2) ;
|
||||||
|
|
||||||
|
% Group 4
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_4', ...
|
||||||
|
'weights', {{init_weight('gaussian', 1, 1, 32, 64*DCTMode, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_4', ...
|
||||||
|
'weights', {{ones(64*DCTMode, 1, 'single'), ...
|
||||||
|
zeros(64*DCTMode, 1, 'single'), ...
|
||||||
|
zeros(64*DCTMode, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'relu', 'name', 'ReLU_4') ;
|
||||||
|
net.layers{end+1} = struct('type', 'pool', ...
|
||||||
|
'name', 'Pool_4', ...
|
||||||
|
'method', 'avg', ...
|
||||||
|
'pool', [5 5], ...
|
||||||
|
'stride', 2, ...
|
||||||
|
'pad', 2) ;
|
||||||
|
|
||||||
|
% Group 5
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_5', ...
|
||||||
|
'weights', {{init_weight('gaussian', 1, 1, 64, 128*DCTMode, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_5', ...
|
||||||
|
'weights', {{ones(128*DCTMode, 1, 'single'), ...
|
||||||
|
zeros(128*DCTMode, 1, 'single'), ...
|
||||||
|
zeros(128*DCTMode, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'relu', 'name', 'ReLU_5') ;
|
||||||
|
net.layers{end+1} = struct('type', 'pool', ...
|
||||||
|
'name', 'Pool_5', ...
|
||||||
|
'method', 'avg', ...
|
||||||
|
'pool', [16 16], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0) ;
|
||||||
|
|
||||||
|
% Full connect layer
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'FC',...
|
||||||
|
'weights', {{init_weight('xavier', 1,1,128*DCTMode,2, 'single'), ...
|
||||||
|
0.01*ones(2, 1, 'single')}}, ...
|
||||||
|
'learningRate', [1 2], ...
|
||||||
|
'weightDecay', [1 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0) ;
|
||||||
|
|
||||||
|
% Softmax layer
|
||||||
|
net.layers{end+1} = struct('type', 'softmaxloss', 'name', 'loss') ;
|
||||||
|
|
||||||
|
% Meta parameters
|
||||||
|
net.meta.inputSize = [512 512 1] ;
|
||||||
|
|
||||||
|
lr = get_lr_sequence(opts.lrSequence);
|
||||||
|
net.meta.trainOpts.learningRate = lr;
|
||||||
|
net.meta.trainOpts.numEpochs = numel(lr) ;
|
||||||
|
net.meta.trainOpts.batchSize = opts.batchSize ;
|
||||||
|
net.meta.trainOpts.weightDecay = 0.01;
|
||||||
|
|
||||||
|
|
||||||
|
% Fill in default values
|
||||||
|
net = vl_simplenn_tidy(net) ;
|
||||||
|
|
||||||
|
% Switch to DagNN if requested
|
||||||
|
switch lower(opts.networkType)
|
||||||
|
case 'simplenn'
|
||||||
|
% done
|
||||||
|
case 'dagnn'
|
||||||
|
net = dagnn.DagNN.fromSimpleNN(net, 'canonicalNames', true) ;
|
||||||
|
net.addLayer('error', dagnn.Loss('loss', 'classerror'), ...
|
||||||
|
{'prediction','label'}, 'error') ;
|
||||||
|
otherwise
|
||||||
|
assert(false) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function weights = init_weight(weightInitMethod, h, w, in, out, type)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% See K. He, X. Zhang, S. Ren, and J. Sun. Delving deep into
|
||||||
|
% rectifiers: Surpassing human-level performance on imagenet
|
||||||
|
% classification. CoRR, (arXiv:1502.01852v1), 2015.
|
||||||
|
switch lower(weightInitMethod)
|
||||||
|
case 'gaussian'
|
||||||
|
sc = 0.01 ;
|
||||||
|
weights = randn(h, w, in, out, type)*sc;
|
||||||
|
case 'xavier'
|
||||||
|
sc = sqrt(3/(h*w*in)) ;
|
||||||
|
weights = (rand(h, w, in, out, type)*2 - 1)*sc ;
|
||||||
|
case 'xavierimproved'
|
||||||
|
sc = sqrt(2/(h*w*out)) ;
|
||||||
|
weights = randn(h, w, in, out, type)*sc ;
|
||||||
|
otherwise
|
||||||
|
error('Unknown weight initialization method''%s''', weightInitMethod);
|
||||||
|
end
|
||||||
|
|
||||||
|
function lr = get_lr_sequence( lrGenerationMethod )
|
||||||
|
|
||||||
|
switch lower(lrGenerationMethod)
|
||||||
|
case 'step_short'
|
||||||
|
lr = 0.001 * ones(1, 2);
|
||||||
|
for i = 1:39
|
||||||
|
lr =[lr, lr(end-1:end)*0.9];
|
||||||
|
end
|
||||||
|
case 'log_short'
|
||||||
|
%lr = logspace(-3, -5, 80);
|
||||||
|
lr = logspace(-3, -5, 40 );
|
||||||
|
case 'step_long'
|
||||||
|
numInterationPerEpoch = 8000/64;
|
||||||
|
lrStepSize = 5000/numInterationPerEpoch; %
|
||||||
|
totalStep = 220000/5000; % CNN is trained for 120,000 iterations
|
||||||
|
lr = 0.001*ones(1,lrStepSize);
|
||||||
|
for i = 1:totalStep - 1
|
||||||
|
lr = [lr, lr(end-lrStepSize+1:end) *0.9];
|
||||||
|
end
|
||||||
|
case 'step_long2'
|
||||||
|
numInterationPerEpoch = 8000/64;
|
||||||
|
lrStepSize = 2500/numInterationPerEpoch; %
|
||||||
|
totalStep = 12;
|
||||||
|
lr = 0.001*ones(1,lrStepSize);
|
||||||
|
for i = 1:totalStep - 1
|
||||||
|
lr = [lr, lr(end-lrStepSize+1:end) *0.75];
|
||||||
|
end
|
||||||
|
case 'step_long3'
|
||||||
|
numInterationPerEpoch = 8000/64;
|
||||||
|
lrStepSize = 2500/numInterationPerEpoch/2; %
|
||||||
|
totalStep = 10;
|
||||||
|
lr = 0.001*ones(1,lrStepSize);
|
||||||
|
for i = 1:totalStep - 1
|
||||||
|
lr = [lr, lr(end-lrStepSize+1:end) *0.5];
|
||||||
|
end
|
||||||
|
otherwise
|
||||||
|
error('unkown type of lr sequence generation method''%s''', lrGenerationMethod);
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,264 @@
|
||||||
|
function net = cnn_phaseaware_VNet_init(varargin)
|
||||||
|
% Define and initialize PhaseAwareNet net
|
||||||
|
opts.networkType = 'dagnn' ;
|
||||||
|
opts.batchSize = 40;
|
||||||
|
opts.seed = 0;
|
||||||
|
opts.lrSequence = 'step_long2';
|
||||||
|
opts = vl_argparse(opts, varargin) ;
|
||||||
|
|
||||||
|
rng( opts.seed );
|
||||||
|
|
||||||
|
net.layers = {} ;
|
||||||
|
|
||||||
|
convOpts = {'CudnnWorkspaceLimit', 1024*1024*1204} ;
|
||||||
|
|
||||||
|
HPF = zeros(5, 5, 1, 4, 'single');
|
||||||
|
HPF(:,:,1,1) = [ -1, 2, -2, 2, -1; ...
|
||||||
|
2, -6, 8, -6, 2; ...
|
||||||
|
-2, 8, -12, 8, -2; ...
|
||||||
|
2, -6, 8, -6, 2; ...
|
||||||
|
-1, 2, -2, 2, -1]/12;
|
||||||
|
|
||||||
|
HPF(:,:,1,2) = [ 0, 0, 5.2, 0, 0; ...
|
||||||
|
0, 23.4, 36.4, 23.4, 0; ...
|
||||||
|
5.2, 36.4, -261.0, 36.4, 5.2; ...
|
||||||
|
0, 23.4, 36.4, 23.4, 0; ...
|
||||||
|
0, 0, 5.2, 0, 0]/261;
|
||||||
|
|
||||||
|
HPF(:,:,1,3) = [ 0.0562, -0.1354, 0.0000, 0.1354, -0.0562; ...
|
||||||
|
0.0818, -0.1970, 0.0000, 0.1970, -0.0818; ...
|
||||||
|
0.0926, -0.2233, 0.0000, 0.2233, -0.0926; ...
|
||||||
|
0.0818, -0.1970, 0.0000, 0.1970, -0.0818; ...
|
||||||
|
0.0562, -0.1354, 0.0000, 0.1354, -0.0562 ];
|
||||||
|
|
||||||
|
HPF(:,:,1,4) = [-0.0562, -0.0818, -0.0926, -0.0818, -0.0562; ...
|
||||||
|
0.1354, 0.1970, 0.2233, 0.1970, 0.1354; ...
|
||||||
|
0.0000, 0.0000, 0.0000, -0.0000, -0.0000; ...
|
||||||
|
-0.1354, -0.1970, -0.2233, -0.1970, -0.1354; ...
|
||||||
|
0.0562, 0.0818, 0.0926, 0.0818, 0.0562 ];
|
||||||
|
|
||||||
|
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'HPFs', ...
|
||||||
|
'weights', {{HPF, []}}, ...
|
||||||
|
'learningRate', [0, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 2, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
|
||||||
|
% Group 1
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_1', ...
|
||||||
|
'weights', {{init_weight('gaussian', 5, 5, 4, 8, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 2, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'abs', 'name', 'ABS_1') ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_1', ...
|
||||||
|
'weights', {{ones(8, 1, 'single'), ...
|
||||||
|
zeros(8, 1, 'single'), ...
|
||||||
|
zeros(8, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'tanh', 'name', 'TanH_1') ;
|
||||||
|
|
||||||
|
|
||||||
|
% Group 2
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_2', ...
|
||||||
|
'weights', {{init_weight('gaussian', 5, 5, 8, 16, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 2, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_2', ...
|
||||||
|
'weights', {{ones(16, 1, 'single'), ...
|
||||||
|
zeros(16, 1, 'single'), ...
|
||||||
|
zeros(16, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'tanh', 'name', 'TanH_2') ;
|
||||||
|
|
||||||
|
|
||||||
|
% Phase split here
|
||||||
|
net.layers{end+1} = struct('type', 'phasesplit', ...
|
||||||
|
'name', 'DCTPhaseSplit', ...
|
||||||
|
'pool', [1, 1], ...
|
||||||
|
'stride', 8, ...
|
||||||
|
'pad', 0 );
|
||||||
|
|
||||||
|
DCTMode = 64;
|
||||||
|
% Group 3
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_3', ...
|
||||||
|
'weights', {{init_weight('gaussian', 1, 1, 16*DCTMode, 128, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_3', ...
|
||||||
|
'weights', {{ones(128, 1, 'single'), ...
|
||||||
|
zeros(128, 1, 'single'), ...
|
||||||
|
zeros(128, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'relu', 'name', 'ReLU_3') ;
|
||||||
|
net.layers{end+1} = struct('type', 'pool', ...
|
||||||
|
'name', 'Pool_3', ...
|
||||||
|
'method', 'avg', ...
|
||||||
|
'pool', [5 5], ...
|
||||||
|
'stride', 2, ...
|
||||||
|
'pad', 2) ;
|
||||||
|
|
||||||
|
% Group 4
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_4', ...
|
||||||
|
'weights', {{init_weight('gaussian', 1, 1, 128, 256, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_4', ...
|
||||||
|
'weights', {{ones(256, 1, 'single'), ...
|
||||||
|
zeros(256, 1, 'single'), ...
|
||||||
|
zeros(256, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'relu', 'name', 'ReLU_4') ;
|
||||||
|
net.layers{end+1} = struct('type', 'pool', ...
|
||||||
|
'name', 'Pool_4', ...
|
||||||
|
'method', 'avg', ...
|
||||||
|
'pool', [5 5], ...
|
||||||
|
'stride', 2, ...
|
||||||
|
'pad', 2) ;
|
||||||
|
|
||||||
|
% Group 5
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'CONV_5', ...
|
||||||
|
'weights', {{init_weight('gaussian', 1, 1, 256, 512, 'single'), ...
|
||||||
|
[]}}, ...
|
||||||
|
'learningRate', [1, 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0, ...
|
||||||
|
'weightDecay', [0, 0], ...
|
||||||
|
'opts', {convOpts}) ;
|
||||||
|
net.layers{end+1} = struct('type', 'bnorm', 'name', 'BN_5', ...
|
||||||
|
'weights', {{ones(512, 1, 'single'), ...
|
||||||
|
zeros(512, 1, 'single'), ...
|
||||||
|
zeros(512, 2, 'single')}}, ...
|
||||||
|
'learningRate', [1 1 0.01], ...
|
||||||
|
'weightDecay', [0 0]) ;
|
||||||
|
net.layers{end+1} = struct('type', 'relu', 'name', 'ReLU_5') ;
|
||||||
|
net.layers{end+1} = struct('type', 'pool', ...
|
||||||
|
'name', 'Pool_5', ...
|
||||||
|
'method', 'avg', ...
|
||||||
|
'pool', [16 16], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0) ;
|
||||||
|
|
||||||
|
% Full connect layer
|
||||||
|
net.layers{end+1} = struct('type', 'conv', ...
|
||||||
|
'name', 'FC',...
|
||||||
|
'weights', {{init_weight('xavier', 1,1,512,2, 'single'), ...
|
||||||
|
0.01*ones(2, 1, 'single')}}, ...
|
||||||
|
'learningRate', [1 2], ...
|
||||||
|
'weightDecay', [1 0], ...
|
||||||
|
'stride', 1, ...
|
||||||
|
'pad', 0) ;
|
||||||
|
|
||||||
|
% Softmax layer
|
||||||
|
net.layers{end+1} = struct('type', 'softmaxloss', 'name', 'loss') ;
|
||||||
|
|
||||||
|
% Meta parameters
|
||||||
|
net.meta.inputSize = [512 512 1] ;
|
||||||
|
|
||||||
|
lr = get_lr_sequence(opts.lrSequence);
|
||||||
|
net.meta.trainOpts.learningRate = lr;
|
||||||
|
net.meta.trainOpts.numEpochs = numel(lr) ;
|
||||||
|
net.meta.trainOpts.batchSize = opts.batchSize ;
|
||||||
|
net.meta.trainOpts.weightDecay = 0.01; % In the paper it is 0.01,
|
||||||
|
%but it is only applied to Batch Normalization
|
||||||
|
|
||||||
|
% Fill in default values
|
||||||
|
net = vl_simplenn_tidy(net) ;
|
||||||
|
|
||||||
|
% Switch to DagNN if requested
|
||||||
|
switch lower(opts.networkType)
|
||||||
|
case 'simplenn'
|
||||||
|
% done
|
||||||
|
case 'dagnn'
|
||||||
|
net = dagnn.DagNN.fromSimpleNN(net, 'canonicalNames', true) ;
|
||||||
|
net.addLayer('error', dagnn.Loss('loss', 'classerror'), ...
|
||||||
|
{'prediction','label'}, 'error') ;
|
||||||
|
otherwise
|
||||||
|
assert(false) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function weights = init_weight(weightInitMethod, h, w, in, out, type)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% See K. He, X. Zhang, S. Ren, and J. Sun. Delving deep into
|
||||||
|
% rectifiers: Surpassing human-level performance on imagenet
|
||||||
|
% classification. CoRR, (arXiv:1502.01852v1), 2015.
|
||||||
|
switch lower(weightInitMethod)
|
||||||
|
case 'gaussian'
|
||||||
|
sc = 0.01 ;
|
||||||
|
weights = randn(h, w, in, out, type)*sc;
|
||||||
|
case 'xavier'
|
||||||
|
sc = sqrt(3/(h*w*in)) ;
|
||||||
|
weights = (rand(h, w, in, out, type)*2 - 1)*sc ;
|
||||||
|
case 'xavierimproved'
|
||||||
|
sc = sqrt(2/(h*w*out)) ;
|
||||||
|
weights = randn(h, w, in, out, type)*sc ;
|
||||||
|
otherwise
|
||||||
|
error('Unknown weight initialization method''%s''', weightInitMethod);
|
||||||
|
end
|
||||||
|
|
||||||
|
function lr = get_lr_sequence( lrGenerationMethod )
|
||||||
|
|
||||||
|
switch lower(lrGenerationMethod)
|
||||||
|
case 'step_short'
|
||||||
|
lr = 0.001 * ones(1, 2);
|
||||||
|
for i = 1:39
|
||||||
|
lr =[lr, lr(end-1:end)*0.9];
|
||||||
|
end
|
||||||
|
case 'log_short'
|
||||||
|
%lr = logspace(-3, -5, 80);
|
||||||
|
lr = logspace(-3, -5, 40 );
|
||||||
|
case 'step_long'
|
||||||
|
numInterationPerEpoch = 8000/64;
|
||||||
|
lrStepSize = 5000/numInterationPerEpoch; %
|
||||||
|
totalStep = 220000/5000; % CNN is trained for 120,000 iterations
|
||||||
|
lr = 0.001*ones(1,lrStepSize);
|
||||||
|
for i = 1:totalStep - 1
|
||||||
|
lr = [lr, lr(end-lrStepSize+1:end) *0.9];
|
||||||
|
end
|
||||||
|
case 'step_long2'
|
||||||
|
numInterationPerEpoch = 8000/64;
|
||||||
|
lrStepSize = 2500/numInterationPerEpoch; %
|
||||||
|
totalStep = 12; % 8: 160 epoch 12: 240 epoch
|
||||||
|
lr = 0.001*ones(1,lrStepSize);
|
||||||
|
for i = 1:totalStep - 1
|
||||||
|
lr = [lr, lr(end-lrStepSize+1:end) *0.75];
|
||||||
|
end
|
||||||
|
case 'step_long3'
|
||||||
|
numInterationPerEpoch = 8000/64;
|
||||||
|
lrStepSize = 2500/numInterationPerEpoch/2; %
|
||||||
|
totalStep = 12;
|
||||||
|
lr = 0.001*ones(1,lrStepSize);
|
||||||
|
for i = 1:totalStep - 1
|
||||||
|
lr = [lr, lr(end-lrStepSize+1:end) *0.5];
|
||||||
|
end
|
||||||
|
otherwise
|
||||||
|
error('unkown type of lr sequence generation method''%s''', lrGenerationMethod);
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,149 @@
|
||||||
|
function imdb = cnn_phaseaware_imdb_setup( varargin )
|
||||||
|
|
||||||
|
opts.seed = 0;
|
||||||
|
opts.coverPath = 'C:\DeepLearning\matconvnet-1.0-beta20\data\JStego\75_mat';
|
||||||
|
opts.stegoPath = 'C:\DeepLearning\matconvnet-1.0-beta20\data\JStego\JUNI_0.4_mat';
|
||||||
|
opts.ratio = [0.6, 0.15, 0.25]; % train, validation, and test
|
||||||
|
opts.libSize = inf;
|
||||||
|
opts = vl_argparse( opts, varargin );
|
||||||
|
|
||||||
|
rng( opts.seed );
|
||||||
|
opts.ratio = opts.ratio/sum(opts.ratio);
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Sanity Check
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
fprintf('sanity check the library images ...') ;
|
||||||
|
targetSize = 10000;
|
||||||
|
expArray = linspace(1, targetSize, targetSize);
|
||||||
|
|
||||||
|
% first, sanilty the two data base
|
||||||
|
list = dir(fullfile(opts.coverPath, '*.mat'));
|
||||||
|
tokens = regexp({list.name}, '([\d]+).mat', 'tokens') ;
|
||||||
|
nameArray = cellfun(@(x) sscanf(x{1}{1}, '%d'), tokens) ;
|
||||||
|
if( ~isequal( sort(nameArray), expArray ) )
|
||||||
|
error('coverPath = %s is corrupted', opts.coverPath);
|
||||||
|
end
|
||||||
|
|
||||||
|
list = dir(fullfile(opts.stegoPath, '*.mat'));
|
||||||
|
tokens = regexp({list.name}, '([\d]+).mat', 'tokens') ;
|
||||||
|
nameArray = cellfun(@(x) sscanf(x{1}{1}, '%d'), tokens) ;
|
||||||
|
if( ~isequal( sort(nameArray), expArray ) )
|
||||||
|
error('stegoPath = %s is corrupted', opts.stegoPath);
|
||||||
|
end
|
||||||
|
fprintf('[checked]\n') ;
|
||||||
|
|
||||||
|
% meta
|
||||||
|
randomImages = randperm( targetSize );
|
||||||
|
|
||||||
|
totalSize = min( opts.libSize, targetSize );
|
||||||
|
|
||||||
|
numTrn = fix( totalSize * opts.ratio(1));
|
||||||
|
numVal = fix( totalSize * opts.ratio(2));
|
||||||
|
numTst = fix( totalSize * opts.ratio(3));
|
||||||
|
|
||||||
|
imdb.classes.name = {'Cover', 'Stego'} ;
|
||||||
|
n = strfind(opts.coverPath, filesep);
|
||||||
|
if( isempty( n ) )
|
||||||
|
coverDes = 'Cover Images';
|
||||||
|
else
|
||||||
|
coverDes = opts.coverPath(n(end)+1:end);
|
||||||
|
end
|
||||||
|
|
||||||
|
n = strfind(opts.stegoPath, filesep);
|
||||||
|
if( isempty( n ) )
|
||||||
|
stegoDes = 'Stego Images';
|
||||||
|
else
|
||||||
|
stegoDes = opts.stegoPath(n(end)+1:end);
|
||||||
|
end
|
||||||
|
|
||||||
|
imdb.classes.description = {coverDes, stegoDes} ;
|
||||||
|
imdb.classes.coverPath = opts.coverPath;
|
||||||
|
imdb.classes.stegoPath = opts.stegoPath;
|
||||||
|
|
||||||
|
fprintf('%d Trn Image, %d Val Images, and %d Test Images \n ', ...
|
||||||
|
numTrn, numVal, numTst) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Training images
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
fprintf('searching training images ...') ;
|
||||||
|
|
||||||
|
names = cell(1, numTrn * 2 );
|
||||||
|
labels = ones(1, numTrn * 2 );
|
||||||
|
for i = 1:numTrn
|
||||||
|
|
||||||
|
idx = randomImages(i);
|
||||||
|
|
||||||
|
names{2*i-1} = fullfile(opts.coverPath, strcat(num2str(idx),'.mat'));
|
||||||
|
labels(2*i - 1) = 1;
|
||||||
|
|
||||||
|
names{2*i} = fullfile(opts.stegoPath, strcat(num2str(idx),'.mat'));
|
||||||
|
labels(2*i) = 2;
|
||||||
|
end
|
||||||
|
|
||||||
|
imdb.images.id = 1:numel(names) ;
|
||||||
|
imdb.images.name = names ;
|
||||||
|
imdb.images.set = ones(1, numel(names)) ;
|
||||||
|
imdb.images.label = labels ;
|
||||||
|
|
||||||
|
fprintf('done\n') ;
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Validation images
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
fprintf('searching validation images ...') ;
|
||||||
|
|
||||||
|
names = cell(1, numVal * 2 );
|
||||||
|
labels = ones(1, numVal * 2 );
|
||||||
|
|
||||||
|
for i = 1:numVal
|
||||||
|
|
||||||
|
idx = randomImages( numTrn + i);
|
||||||
|
|
||||||
|
names{2*i-1} = fullfile(opts.coverPath, strcat(num2str(idx),'.mat'));
|
||||||
|
labels(2*i - 1) = 1;
|
||||||
|
|
||||||
|
names{2*i} = fullfile(opts.stegoPath, strcat(num2str(idx),'.mat'));
|
||||||
|
labels(2*i) = 2;
|
||||||
|
|
||||||
|
end
|
||||||
|
|
||||||
|
imdb.images.id = horzcat( imdb.images.id, (1:numel(names)) + 1e7 - 1 );
|
||||||
|
imdb.images.name = horzcat(imdb.images.name, names );
|
||||||
|
imdb.images.set = horzcat( imdb.images.set, 2 * ones(1, numel(names)));
|
||||||
|
imdb.images.label = horzcat( imdb.images.label, labels ) ;
|
||||||
|
|
||||||
|
fprintf('done\n') ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Test images
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
fprintf('searching test images ...') ;
|
||||||
|
|
||||||
|
names = cell(1, numTst * 2 );
|
||||||
|
labels = ones(1, numTst * 2 );
|
||||||
|
|
||||||
|
for i = 1:numTst
|
||||||
|
|
||||||
|
idx = randomImages( numTrn + numVal + i);
|
||||||
|
|
||||||
|
names{2*i-1} = fullfile(opts.coverPath, strcat(num2str(idx),'.mat'));
|
||||||
|
labels(2*i - 1) = 1;
|
||||||
|
|
||||||
|
names{2*i} = fullfile(opts.stegoPath, strcat(num2str(idx),'.mat'));
|
||||||
|
labels(2*i) = 2;
|
||||||
|
|
||||||
|
end
|
||||||
|
|
||||||
|
imdb.images.id = horzcat( imdb.images.id, (1:numel(names)) + 2e7 - 1 );
|
||||||
|
imdb.images.name = horzcat(imdb.images.name, names );
|
||||||
|
imdb.images.set = horzcat( imdb.images.set, 3 * ones(1, numel(names)));
|
||||||
|
imdb.images.label = horzcat( imdb.images.label, labels ) ;
|
||||||
|
|
||||||
|
fprintf('done\n') ;
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
@ -0,0 +1,72 @@
|
||||||
|
function [stats, state] = test_phaseaware(varargin)
|
||||||
|
% Test the phasesplit net
|
||||||
|
|
||||||
|
opts.batchSize = 40;
|
||||||
|
opts.expDir = fullfile('data', 'JUNI-7504-PNet-dagnn-40-Seed-0-log_short') ;
|
||||||
|
opts.testEpoch = 40;
|
||||||
|
opts.testSelect = [0, 1, 1]; % (1) training; (2)validation; (3), testing
|
||||||
|
opts.saveResult = true;
|
||||||
|
opts.bnRefine = true;
|
||||||
|
|
||||||
|
opts = vl_argparse( opts, varargin );
|
||||||
|
|
||||||
|
opts.imdbPath = fullfile(opts.expDir, 'imdb.mat');
|
||||||
|
|
||||||
|
opts.train = struct('gpus', [1, 2], 'cudnn', true, 'stegoShuffle', false ) ;
|
||||||
|
%opts.train = struct('gpus', [], 'stegoShuffle', true) ; // CPU debugging
|
||||||
|
if ~isfield(opts.train, 'gpus'), opts.train.gpus = []; end;
|
||||||
|
|
||||||
|
% put it to drawing
|
||||||
|
if ( ~exist( opts.expDir, 'dir' ) )
|
||||||
|
error('expDir is empty' );
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Find the data base
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
if exist(opts.imdbPath, 'file')
|
||||||
|
imdb = load(opts.imdbPath) ;
|
||||||
|
else
|
||||||
|
error(' cannot find imdb' );
|
||||||
|
end
|
||||||
|
|
||||||
|
meta.inputSize = [512, 512, 1, opts.batchSize];
|
||||||
|
|
||||||
|
[state, stats] = cnn_test_dag(imdb, getBatchFn( opts, meta ), ...
|
||||||
|
'expDir', opts.expDir, ...
|
||||||
|
'batchSize', opts.batchSize, ...
|
||||||
|
'testEpoch', opts.testEpoch, ...
|
||||||
|
'testSelect', opts.testSelect, ...
|
||||||
|
'saveResult', opts.saveResult, ...
|
||||||
|
'bnRefine', opts.bnRefine, ...
|
||||||
|
opts.train ) ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function fn = getBatchFn(opts, meta)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
bopts.useGpu = numel(opts.train.gpus) > 0 ;
|
||||||
|
bopts.imageSize = meta.inputSize;
|
||||||
|
|
||||||
|
fn = @(x,y) getDagNNBatch(bopts,x,y) ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function inputs = getDagNNBatch(opts, imdb, batch)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% label
|
||||||
|
labels = imdb.images.label(1,batch) ;
|
||||||
|
% images
|
||||||
|
images = zeros(opts.imageSize(1), opts.imageSize(2), ...
|
||||||
|
opts.imageSize(3), numel(batch), 'single') ;
|
||||||
|
for i = 1:numel(batch)
|
||||||
|
% imt = imread(imdb.images.name{batch(i)});
|
||||||
|
% images(:,:,:,i) = single(imt);
|
||||||
|
imt = load(imdb.images.name{batch(i)}, 'im');
|
||||||
|
images(:,:,:,i) = single(imt.im);
|
||||||
|
end
|
||||||
|
|
||||||
|
if opts.useGpu > 0
|
||||||
|
images = gpuArray(images) ;
|
||||||
|
end
|
||||||
|
inputs = {'input', images, 'label', labels} ;
|
||||||
|
|
@ -0,0 +1,284 @@
|
||||||
|
function [BN_Moments,stats] = cnn_bnrefine_dag( imdb, getBatch, varargin)
|
||||||
|
%CNN_TEST_DAG Demonstrates test a CNN using the DagNN wrapper
|
||||||
|
% CNN_TEST_DAG() is a slim version to CNN_TRAIN_DAG(), just do the
|
||||||
|
% testing of the final net in the export
|
||||||
|
|
||||||
|
opts.expDir = fullfile('data','exp') ;
|
||||||
|
opts.batchSize = 256 ;
|
||||||
|
opts.train = [] ;
|
||||||
|
opts.val = [] ;
|
||||||
|
opts.test = [];
|
||||||
|
opts.gpus = [] ;
|
||||||
|
opts.prefetch = false ;
|
||||||
|
opts.testEpoch = inf;
|
||||||
|
opts.bnEpochCollectSize = 2000;
|
||||||
|
opts.saveResult = true;
|
||||||
|
|
||||||
|
opts.randomSeed = 0 ;
|
||||||
|
opts.stegoShuffle = false;
|
||||||
|
opts.cudnn = true ;
|
||||||
|
opts.extractStatsFn = @extractStats ;
|
||||||
|
|
||||||
|
opts = vl_argparse(opts, varargin) ;
|
||||||
|
|
||||||
|
if ~exist(opts.expDir, 'dir'), mkdir(opts.expDir) ; end
|
||||||
|
if isempty(opts.train), opts.train = find(imdb.images.set==1) ; end
|
||||||
|
if isnan(opts.train), opts.train = [] ; end
|
||||||
|
|
||||||
|
% we must restrict the BN moment pooling from train set only
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Initialization
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
state.getBatch = getBatch ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Train and validate
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
modelPath = @(ep) fullfile(opts.expDir, sprintf('net-epoch-%d.mat', ep));
|
||||||
|
resultPath = @(ep) fullfile(opts.expDir, sprintf('bn-epoch-%d.mat', ep));
|
||||||
|
|
||||||
|
start = findLastCheckpoint(opts.expDir) ;
|
||||||
|
if( start < 1 )
|
||||||
|
error( 'Found no net' );
|
||||||
|
end
|
||||||
|
|
||||||
|
if start >= 1
|
||||||
|
start = min(start, opts.testEpoch);
|
||||||
|
fprintf('%s: testing by loading epoch %d\n', mfilename, start) ;
|
||||||
|
net = loadState(modelPath(start)) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% First, create the structure to pool the BN moments
|
||||||
|
numLayers = numel(net.layers);
|
||||||
|
|
||||||
|
BN_Moments = struct('layer', {}, ...
|
||||||
|
'name', {}, ...
|
||||||
|
'inputs', {}, ...
|
||||||
|
'outputs', {}, ...
|
||||||
|
'shape', {}, ...
|
||||||
|
'dataType', {}, ...
|
||||||
|
'oldValue', {}, ...
|
||||||
|
'hist', {} ) ;
|
||||||
|
|
||||||
|
for i = 1:numLayers
|
||||||
|
if ( isa( net.layers(i).block, 'dagnn.BatchNorm') )
|
||||||
|
% Neet to save the BN moments for pooling
|
||||||
|
net.layers(i).block.computeMoment = true;
|
||||||
|
|
||||||
|
name = net.layers(i).params{3};
|
||||||
|
dataType = class(net.getParam(name).value);
|
||||||
|
shape = size(net.getParam(name).value);
|
||||||
|
|
||||||
|
BN_Moments(end+1).layer = net.layers(i).name;
|
||||||
|
BN_Moments(end).name = name ;
|
||||||
|
BN_Moments(end).inputs = net.layers(i).inputs;
|
||||||
|
BN_Moments(end).outputs = net.layers(i).outputs;
|
||||||
|
BN_Moments(end).shape = shape ;
|
||||||
|
BN_Moments(end).dataType = dataType ;
|
||||||
|
BN_Moments(end).oldValue = net.getParam(name).value;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
|
||||||
|
if( numel(opts.gpus) > 1 )
|
||||||
|
error( 'cannot support multiple GPU now ')
|
||||||
|
end
|
||||||
|
|
||||||
|
numEpoch = ceil(opts.bnEpochCollectSize/(numel(opts.train)/opts.batchSize));
|
||||||
|
|
||||||
|
rng(start + opts.randomSeed) ;
|
||||||
|
|
||||||
|
for epoch = start:start + numEpoch - 1
|
||||||
|
|
||||||
|
% Set the random seed based on the epoch and opts.randomSeed.
|
||||||
|
% This is important for reproducibility, including when training
|
||||||
|
% is restarted from a checkpoint.
|
||||||
|
|
||||||
|
prepareGPUs( opts, true ) ;
|
||||||
|
|
||||||
|
% Train for one epoch.
|
||||||
|
state.epoch = epoch ;
|
||||||
|
|
||||||
|
% shuffle
|
||||||
|
if( opts.stegoShuffle )
|
||||||
|
|
||||||
|
N = numel(opts.train); % TRN
|
||||||
|
|
||||||
|
Lab = max( 1, numel(opts.gpus));
|
||||||
|
|
||||||
|
% M and N must be even, and multiple Lab
|
||||||
|
assert( rem( N, 2*Lab ) == 0 );
|
||||||
|
|
||||||
|
seq = opts.train( 2*randperm(N/2) - 1 );
|
||||||
|
seq = reshape( seq, Lab, N/(2*Lab) );
|
||||||
|
state.train = reshape( [seq; seq+1], 1, N );
|
||||||
|
|
||||||
|
else
|
||||||
|
|
||||||
|
state.train = opts.train(randperm(numel(opts.train))) ;
|
||||||
|
|
||||||
|
end
|
||||||
|
|
||||||
|
state.imdb = imdb ;
|
||||||
|
|
||||||
|
% keep pooling the result
|
||||||
|
[stats.train(epoch - start + 1), BN_Moments] = process_epoch(net, state, opts, BN_Moments ) ;
|
||||||
|
|
||||||
|
end
|
||||||
|
|
||||||
|
% Reset the parameters
|
||||||
|
for i = 1:numel(BN_Moments)
|
||||||
|
bn_moment_name = BN_Moments(i).name;
|
||||||
|
statsVal = median(BN_Moments(i).hist, 3);
|
||||||
|
|
||||||
|
% set the new value
|
||||||
|
paramIdx = net.getParamIndex(bn_moment_name);
|
||||||
|
% double check the shape, see if it matches
|
||||||
|
assert( isequal(size(statsVal), size(net.params(paramIdx).value ) ) );
|
||||||
|
|
||||||
|
% reset the BN moment parameters
|
||||||
|
net.params(paramIdx).value = statsVal;
|
||||||
|
end
|
||||||
|
|
||||||
|
% Revert it back
|
||||||
|
for i = 1:numel(net.layers)
|
||||||
|
if ( isa( net.layers(i).block, 'dagnn.BatchNorm') )
|
||||||
|
net.layers(i).block.computeMoment = false;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
|
||||||
|
saveState(resultPath(start), net, stats, BN_Moments ) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function [stats, BN_Moments] = process_epoch(net, state, opts, BN_Moments )
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
% move CNN to GPU as needed
|
||||||
|
numGpus = numel(opts.gpus) ;
|
||||||
|
if numGpus >= 1
|
||||||
|
net.move('gpu') ;
|
||||||
|
end
|
||||||
|
|
||||||
|
subset = state.train;
|
||||||
|
num = 0 ;
|
||||||
|
stats.num = 0 ; % return something even if subset = []
|
||||||
|
stats.time = 0 ;
|
||||||
|
adjustTime = 0 ;
|
||||||
|
|
||||||
|
start = tic ;
|
||||||
|
for t=1:opts.batchSize:numel(subset)
|
||||||
|
fprintf('%s: epoch %02d: %3d/%3d:', 'test', state.epoch, ...
|
||||||
|
fix((t-1)/opts.batchSize)+1, ceil(numel(subset)/opts.batchSize)) ;
|
||||||
|
batchSize = min(opts.batchSize, numel(subset) - t + 1) ;
|
||||||
|
|
||||||
|
% get this image batch and prefetch the next
|
||||||
|
s = 1;
|
||||||
|
batchStart = t + (labindex-1) + (s-1) * numlabs ;
|
||||||
|
batchEnd = min(t+opts.batchSize-1, numel(subset)) ;
|
||||||
|
batch = subset(batchStart : numlabs : batchEnd) ;
|
||||||
|
num = num + numel(batch) ;
|
||||||
|
if numel(batch) == 0, continue ; end
|
||||||
|
|
||||||
|
inputs = state.getBatch(state.imdb, batch) ;
|
||||||
|
|
||||||
|
net.mode = 'test' ;
|
||||||
|
|
||||||
|
net.eval(inputs) ;
|
||||||
|
|
||||||
|
% update here
|
||||||
|
for i = 1:numel(BN_Moments)
|
||||||
|
layer_name = BN_Moments(i).layer;
|
||||||
|
newVal = gather( net.getLayer(layer_name).block.moments );
|
||||||
|
assert( ~isempty( newVal ) ); % in case the BatchNorm is not set up
|
||||||
|
BN_Moments(i).hist = cat( 3, BN_Moments(i).hist, newVal );
|
||||||
|
end
|
||||||
|
|
||||||
|
|
||||||
|
% get statistics
|
||||||
|
time = toc(start) + adjustTime ;
|
||||||
|
batchTime = time - stats.time ;
|
||||||
|
stats = opts.extractStatsFn(net) ;
|
||||||
|
stats.num = num ;
|
||||||
|
stats.time = time ;
|
||||||
|
currentSpeed = batchSize / batchTime ;
|
||||||
|
averageSpeed = (t + batchSize - 1) / time ;
|
||||||
|
if t == opts.batchSize + 1
|
||||||
|
% compensate for the first iteration, which is an outlier
|
||||||
|
adjustTime = 2*batchTime - time ;
|
||||||
|
stats.time = time + adjustTime ;
|
||||||
|
end
|
||||||
|
|
||||||
|
fprintf(' %.1f (%.1f) Hz', averageSpeed, currentSpeed) ;
|
||||||
|
for f = setdiff(fieldnames(stats)', {'num', 'time'})
|
||||||
|
f = char(f) ;
|
||||||
|
fprintf(' %s:', f) ;
|
||||||
|
fprintf(' %.3f', stats.(f)) ;
|
||||||
|
end
|
||||||
|
fprintf('\n') ;
|
||||||
|
end
|
||||||
|
|
||||||
|
net.reset() ;
|
||||||
|
net.move('cpu') ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function stats = extractStats(net)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
sel = find(cellfun(@(x) isa(x,'dagnn.Loss'), {net.layers.block})) ;
|
||||||
|
stats = struct() ;
|
||||||
|
for i = 1:numel(sel)
|
||||||
|
stats.(net.layers(sel(i)).outputs{1}) = net.layers(sel(i)).block.average ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function saveState(fileName, net, stats, BN_Moments )
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
net_ = net ;
|
||||||
|
net = net_.saveobj() ;
|
||||||
|
save(fileName, 'net', 'stats', 'BN_Moments') ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function net = loadState(fileName)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
load(fileName, 'net' ) ;
|
||||||
|
net = dagnn.DagNN.loadobj(net) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function epoch = findLastCheckpoint(modelDir)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
list = dir(fullfile(modelDir, 'net-epoch-*.mat')) ;
|
||||||
|
tokens = regexp({list.name}, 'net-epoch-([\d]+).mat', 'tokens') ;
|
||||||
|
epoch = cellfun(@(x) sscanf(x{1}{1}, '%d'), tokens) ;
|
||||||
|
epoch = max([epoch 0]) ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function prepareGPUs(opts, cold)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
numGpus = numel(opts.gpus) ;
|
||||||
|
if numGpus > 1
|
||||||
|
% check parallel pool integrity as it could have timed out
|
||||||
|
pool = gcp('nocreate') ;
|
||||||
|
if ~isempty(pool) && pool.NumWorkers ~= numGpus
|
||||||
|
delete(pool) ;
|
||||||
|
end
|
||||||
|
pool = gcp('nocreate') ;
|
||||||
|
if isempty(pool)
|
||||||
|
parpool('local', numGpus) ;
|
||||||
|
cold = true ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
if numGpus >= 1 && cold
|
||||||
|
fprintf('%s: resetting GPU\n', mfilename)
|
||||||
|
if numGpus == 1
|
||||||
|
gpuDevice(opts.gpus)
|
||||||
|
else
|
||||||
|
spmd, gpuDevice(opts.gpus(labindex)), end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
%end
|
||||||
|
|
@ -0,0 +1,341 @@
|
||||||
|
function [state,stats] = cnn_test_dag( imdb, getBatch, varargin)
|
||||||
|
%CNN_TEST_DAG Demonstrates test a CNN using the DagNN wrapper
|
||||||
|
% CNN_TEST_DAG() is a slim version to CNN_TRAIN_DAG(), just do the
|
||||||
|
% testing of the final net in the export
|
||||||
|
|
||||||
|
opts.expDir = fullfile('data','exp') ;
|
||||||
|
opts.batchSize = 256 ;
|
||||||
|
opts.numSubBatches = 1 ;
|
||||||
|
opts.train = [] ;
|
||||||
|
opts.val = [] ;
|
||||||
|
opts.test = [];
|
||||||
|
opts.gpus = [] ;
|
||||||
|
opts.prefetch = false ;
|
||||||
|
opts.testEpoch = inf;
|
||||||
|
opts.testSelect = [1, 1, 1]; % (1) training; (2)validation; (3), testing
|
||||||
|
opts.saveResult = true;
|
||||||
|
opts.bnRefine = false;
|
||||||
|
|
||||||
|
opts.randomSeed = 0 ;
|
||||||
|
opts.stegoShuffle = false;
|
||||||
|
opts.cudnn = true ;
|
||||||
|
opts.extractStatsFn = @extractStats ;
|
||||||
|
|
||||||
|
opts = vl_argparse(opts, varargin) ;
|
||||||
|
|
||||||
|
if ~exist(opts.expDir, 'dir'), mkdir(opts.expDir) ; end
|
||||||
|
if isempty(opts.train), opts.train = find(imdb.images.set==1) ; end
|
||||||
|
if isempty(opts.val), opts.val = find(imdb.images.set==2) ; end
|
||||||
|
if isempty(opts.test), opts.test = find(imdb.images.set==3); end
|
||||||
|
if isnan(opts.train), opts.train = [] ; end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Initialization
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
state.getBatch = getBatch ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Train and validate
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
if ( opts.bnRefine )
|
||||||
|
modelPath = @(ep) fullfile(opts.expDir, sprintf('bn-epoch-%d.mat', ep));
|
||||||
|
resultPath = @(ep) fullfile(opts.expDir, sprintf('test-bn-epoch-%d.mat', ep));
|
||||||
|
else
|
||||||
|
modelPath = @(ep) fullfile(opts.expDir, sprintf('net-epoch-%d.mat', ep));
|
||||||
|
resultPath = @(ep) fullfile(opts.expDir, sprintf('test-net-epoch-%d.mat', ep));
|
||||||
|
end
|
||||||
|
|
||||||
|
start = findLastCheckpoint(opts.expDir) ;
|
||||||
|
if( start < 1 )
|
||||||
|
error( 'Found no net' );
|
||||||
|
end
|
||||||
|
|
||||||
|
if start >= 1
|
||||||
|
start = min(start, opts.testEpoch);
|
||||||
|
fprintf('%s: testing by loading epoch name %s\n', mfilename, modelPath(start) );
|
||||||
|
net = loadState(modelPath(start)) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% Make sure that we use the estimated BN moments
|
||||||
|
for i = 1:numel(net.layers)
|
||||||
|
if ( isa( net.layers(i).block, 'dagnn.BatchNorm') )
|
||||||
|
net.layers(i).block.computeMoment = false;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
|
||||||
|
for epoch = start
|
||||||
|
|
||||||
|
% Set the random seed based on the epoch and opts.randomSeed.
|
||||||
|
% This is important for reproducibility, including when training
|
||||||
|
% is restarted from a checkpoint.
|
||||||
|
|
||||||
|
rng(epoch + opts.randomSeed) ;
|
||||||
|
prepareGPUs(opts, true ) ;
|
||||||
|
|
||||||
|
% Train for one epoch.
|
||||||
|
state.epoch = epoch ;
|
||||||
|
|
||||||
|
% shuffle
|
||||||
|
if( opts.stegoShuffle )
|
||||||
|
|
||||||
|
N = numel(opts.train); % TRN
|
||||||
|
M = numel(opts.val); % VAL
|
||||||
|
K = numel(opts.test); % TST
|
||||||
|
|
||||||
|
Lab = max( 1, numel(opts.gpus));
|
||||||
|
|
||||||
|
% M and N must be even, and multiple Lab
|
||||||
|
assert( ( rem( N, 2*Lab ) == 0 ) & ...
|
||||||
|
( rem( M, 2*Lab ) == 0 ) & ...
|
||||||
|
( rem( K, 2*Lab ) == 0 ) );
|
||||||
|
|
||||||
|
seq = opts.train( 2*randperm(N/2) - 1 );
|
||||||
|
seq = reshape( seq, Lab, N/(2*Lab) );
|
||||||
|
state.train = reshape( [seq; seq+1], 1, N );
|
||||||
|
|
||||||
|
seq = opts.val( 2*randperm(M/2) - 1 );
|
||||||
|
seq = reshape( seq, Lab, M/(2*Lab) );
|
||||||
|
state.val = reshape( [seq; seq+1], 1, M );
|
||||||
|
|
||||||
|
seq = opts.test( 2*randperm(K/2) - 1 );
|
||||||
|
seq = reshape( seq, Lab, K/(2*Lab) );
|
||||||
|
state.test = reshape( [seq; seq+1], 1, K );
|
||||||
|
|
||||||
|
else
|
||||||
|
|
||||||
|
state.train = opts.train(randperm(numel(opts.train))) ;
|
||||||
|
state.val = opts.val(randperm(numel(opts.val))) ;
|
||||||
|
state.test = opts.test(randperm(numel(opts.test))) ;
|
||||||
|
|
||||||
|
% N = numel(opts.train); % TRN
|
||||||
|
% M = numel(opts.val); % VAL
|
||||||
|
% K = numel(opts.test); % TST
|
||||||
|
%
|
||||||
|
%
|
||||||
|
% state.train = opts.train([1:2:N, 2:2:N]);
|
||||||
|
% state.val = opts.val([1:2:M, 2:2:M]);
|
||||||
|
% state.test = opts.test([1:2:K, 2:2:K]);
|
||||||
|
|
||||||
|
end
|
||||||
|
|
||||||
|
state.imdb = imdb ;
|
||||||
|
|
||||||
|
if numel(opts.gpus) <= 1
|
||||||
|
if( opts.testSelect(1) )
|
||||||
|
stats.train = process_epoch(net, state, opts, 'train') ;
|
||||||
|
end
|
||||||
|
if( opts.testSelect(2) )
|
||||||
|
stats.val = process_epoch(net, state, opts, 'val') ;
|
||||||
|
end
|
||||||
|
if( opts.testSelect(3) )
|
||||||
|
stats.test = process_epoch(net, state, opts, 'test');
|
||||||
|
end
|
||||||
|
|
||||||
|
else
|
||||||
|
savedNet = net.saveobj() ;
|
||||||
|
spmd
|
||||||
|
net_ = dagnn.DagNN.loadobj(savedNet) ;
|
||||||
|
if( opts.testSelect(1) )
|
||||||
|
stats_.train = process_epoch(net_, state, opts, 'train') ;
|
||||||
|
end
|
||||||
|
if( opts.testSelect(2) )
|
||||||
|
stats_.val = process_epoch(net_, state, opts, 'val') ;
|
||||||
|
end
|
||||||
|
if( opts.testSelect(3) )
|
||||||
|
stats_.test = process_epoch(net_, state, opts, 'test');
|
||||||
|
end
|
||||||
|
if labindex == 1, savedNet_ = net_.saveobj() ; end
|
||||||
|
end
|
||||||
|
net = dagnn.DagNN.loadobj(savedNet_{1}) ;
|
||||||
|
stats__ = accumulateStats(stats_) ;
|
||||||
|
|
||||||
|
if( opts.testSelect(1) )
|
||||||
|
stats.train = stats__.train ;
|
||||||
|
end
|
||||||
|
if( opts.testSelect(2) )
|
||||||
|
stats.val = stats__.val ;
|
||||||
|
end
|
||||||
|
if( opts.testSelect(3) )
|
||||||
|
stats.test = stats__.test;
|
||||||
|
end
|
||||||
|
|
||||||
|
clear net_ stats_ stats__ savedNet savedNet_ ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% save
|
||||||
|
if( opts.saveResult == true )
|
||||||
|
saveState(resultPath(epoch), net, stats, state) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function stats = process_epoch(net, state, opts, mode)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
% move CNN to GPU as needed
|
||||||
|
numGpus = numel(opts.gpus) ;
|
||||||
|
if numGpus >= 1
|
||||||
|
net.move('gpu') ;
|
||||||
|
end
|
||||||
|
|
||||||
|
subset = state.(mode) ;
|
||||||
|
num = 0 ;
|
||||||
|
stats.num = 0 ; % return something even if subset = []
|
||||||
|
stats.time = 0 ;
|
||||||
|
adjustTime = 0 ;
|
||||||
|
|
||||||
|
start = tic ;
|
||||||
|
for t=1:opts.batchSize:numel(subset)
|
||||||
|
fprintf('%s: epoch %02d: %3d/%3d:', mode, state.epoch, ...
|
||||||
|
fix((t-1)/opts.batchSize)+1, ceil(numel(subset)/opts.batchSize)) ;
|
||||||
|
batchSize = min(opts.batchSize, numel(subset) - t + 1) ;
|
||||||
|
|
||||||
|
for s=1:opts.numSubBatches
|
||||||
|
% get this image batch and prefetch the next
|
||||||
|
batchStart = t + (labindex-1) + (s-1) * numlabs ;
|
||||||
|
batchEnd = min(t+opts.batchSize-1, numel(subset)) ;
|
||||||
|
batch = subset(batchStart : opts.numSubBatches * numlabs : batchEnd) ;
|
||||||
|
num = num + numel(batch) ;
|
||||||
|
if numel(batch) == 0, continue ; end
|
||||||
|
|
||||||
|
inputs = state.getBatch(state.imdb, batch) ;
|
||||||
|
|
||||||
|
if opts.prefetch
|
||||||
|
if s == opts.numSubBatches
|
||||||
|
batchStart = t + (labindex-1) + opts.batchSize ;
|
||||||
|
batchEnd = min(t+2*opts.batchSize-1, numel(subset)) ;
|
||||||
|
else
|
||||||
|
batchStart = batchStart + numlabs ;
|
||||||
|
end
|
||||||
|
nextBatch = subset(batchStart : opts.numSubBatches * numlabs : batchEnd) ;
|
||||||
|
state.getBatch(state.imdb, nextBatch) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
net.mode = 'test' ;
|
||||||
|
net.eval(inputs) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% get statistics
|
||||||
|
time = toc(start) + adjustTime ;
|
||||||
|
batchTime = time - stats.time ;
|
||||||
|
stats = opts.extractStatsFn(net) ;
|
||||||
|
stats.num = num ;
|
||||||
|
stats.time = time ;
|
||||||
|
currentSpeed = batchSize / batchTime ;
|
||||||
|
averageSpeed = (t + batchSize - 1) / time ;
|
||||||
|
if t == opts.batchSize + 1
|
||||||
|
% compensate for the first iteration, which is an outlier
|
||||||
|
adjustTime = 2*batchTime - time ;
|
||||||
|
stats.time = time + adjustTime ;
|
||||||
|
end
|
||||||
|
|
||||||
|
fprintf(' %.1f (%.1f) Hz', averageSpeed, currentSpeed) ;
|
||||||
|
for f = setdiff(fieldnames(stats)', {'num', 'time'})
|
||||||
|
f = char(f) ;
|
||||||
|
fprintf(' %s:', f) ;
|
||||||
|
fprintf(' %.3f', stats.(f)) ;
|
||||||
|
end
|
||||||
|
fprintf('\n') ;
|
||||||
|
end
|
||||||
|
|
||||||
|
net.reset() ;
|
||||||
|
net.move('cpu') ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function stats = accumulateStats(stats_)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
for s = {'train', 'val', 'test'}
|
||||||
|
s = char(s) ;
|
||||||
|
total = 0 ;
|
||||||
|
|
||||||
|
% initialize stats stucture with same fields and same order as
|
||||||
|
% stats_{1}
|
||||||
|
stats__ = stats_{1} ;
|
||||||
|
if ( ~isfield(stats__, s) )
|
||||||
|
continue;
|
||||||
|
end
|
||||||
|
names = fieldnames(stats__.(s))' ;
|
||||||
|
values = zeros(1, numel(names)) ;
|
||||||
|
fields = cat(1, names, num2cell(values)) ;
|
||||||
|
stats.(s) = struct(fields{:}) ;
|
||||||
|
|
||||||
|
for g = 1:numel(stats_)
|
||||||
|
stats__ = stats_{g} ;
|
||||||
|
num__ = stats__.(s).num ;
|
||||||
|
total = total + num__ ;
|
||||||
|
|
||||||
|
for f = setdiff(fieldnames(stats__.(s))', 'num')
|
||||||
|
f = char(f) ;
|
||||||
|
stats.(s).(f) = stats.(s).(f) + stats__.(s).(f) * num__ ;
|
||||||
|
|
||||||
|
if g == numel(stats_)
|
||||||
|
stats.(s).(f) = stats.(s).(f) / total ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
stats.(s).num = total ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function stats = extractStats(net)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
sel = find(cellfun(@(x) isa(x,'dagnn.Loss'), {net.layers.block})) ;
|
||||||
|
stats = struct() ;
|
||||||
|
for i = 1:numel(sel)
|
||||||
|
stats.(net.layers(sel(i)).outputs{1}) = net.layers(sel(i)).block.average ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function saveState(fileName, net, stats, state )
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
net_ = net ;
|
||||||
|
net = net_.saveobj() ;
|
||||||
|
save(fileName, 'net', 'stats', 'state') ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function [net, stats] = loadState(fileName)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
load(fileName, 'net', 'stats') ;
|
||||||
|
net = dagnn.DagNN.loadobj(net) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function epoch = findLastCheckpoint(modelDir)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
list = dir(fullfile(modelDir, 'net-epoch-*.mat')) ;
|
||||||
|
tokens = regexp({list.name}, 'net-epoch-([\d]+).mat', 'tokens') ;
|
||||||
|
epoch = cellfun(@(x) sscanf(x{1}{1}, '%d'), tokens) ;
|
||||||
|
epoch = max([epoch 0]) ;
|
||||||
|
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function prepareGPUs(opts, cold)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
numGpus = numel(opts.gpus) ;
|
||||||
|
if numGpus > 1
|
||||||
|
% check parallel pool integrity as it could have timed out
|
||||||
|
pool = gcp('nocreate') ;
|
||||||
|
if ~isempty(pool) && pool.NumWorkers ~= numGpus
|
||||||
|
delete(pool) ;
|
||||||
|
end
|
||||||
|
pool = gcp('nocreate') ;
|
||||||
|
if isempty(pool)
|
||||||
|
parpool('local', numGpus) ;
|
||||||
|
cold = true ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
if numGpus >= 1 && cold
|
||||||
|
fprintf('%s: resetting GPU\n', mfilename)
|
||||||
|
if numGpus == 1
|
||||||
|
gpuDevice(opts.gpus)
|
||||||
|
else
|
||||||
|
spmd, gpuDevice(opts.gpus(labindex)), end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
%end
|
||||||
|
|
@ -0,0 +1,516 @@
|
||||||
|
function [net,stats] = cnn_train_dag(net, imdb, getBatch, varargin)
|
||||||
|
%CNN_TRAIN_DAG Demonstrates training a CNN using the DagNN wrapper
|
||||||
|
% CNN_TRAIN_DAG() is similar to CNN_TRAIN(), but works with
|
||||||
|
% the DagNN wrapper instead of the SimpleNN wrapper.
|
||||||
|
|
||||||
|
% Copyright (C) 2014-16 Andrea Vedaldi.
|
||||||
|
% All rights reserved.
|
||||||
|
%
|
||||||
|
% This file is part of the VLFeat library and is made available under
|
||||||
|
% the terms of the BSD license (see the COPYING file).
|
||||||
|
|
||||||
|
opts.expDir = fullfile('data','exp') ;
|
||||||
|
opts.continue = true ;
|
||||||
|
opts.batchSize = 256 ;
|
||||||
|
opts.numSubBatches = 1 ;
|
||||||
|
opts.train = [] ;
|
||||||
|
opts.val = [] ;
|
||||||
|
opts.gpus = [] ;
|
||||||
|
opts.prefetch = false ;
|
||||||
|
opts.numEpochs = 300 ;
|
||||||
|
opts.learningRate = 0.001 ;
|
||||||
|
opts.weightDecay = 0.0005 ;
|
||||||
|
opts.momentum = 0.9 ;
|
||||||
|
opts.randomSeed = 0 ;
|
||||||
|
opts.stegoShuffle = false;
|
||||||
|
opts.computeBNMoment = false;
|
||||||
|
opts.memoryMapFile = fullfile(tempdir, 'matconvnet.bin') ;
|
||||||
|
opts.profile = false ;
|
||||||
|
opts.cudnn = true ;
|
||||||
|
|
||||||
|
opts.derOutputs = {'objective', 1} ;
|
||||||
|
opts.extractStatsFn = @extractStats ;
|
||||||
|
opts.plotStatistics = true;
|
||||||
|
opts = vl_argparse(opts, varargin) ;
|
||||||
|
|
||||||
|
if ~exist(opts.expDir, 'dir'), mkdir(opts.expDir) ; end
|
||||||
|
if isempty(opts.train), opts.train = find(imdb.images.set==1) ; end
|
||||||
|
if isempty(opts.val), opts.val = find(imdb.images.set==2) ; end
|
||||||
|
if isnan(opts.train), opts.train = [] ; end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Initialization
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
evaluateMode = isempty(opts.train) ;
|
||||||
|
if ~evaluateMode
|
||||||
|
if isempty(opts.derOutputs)
|
||||||
|
error('DEROUTPUTS must be specified when training.\n') ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
state.getBatch = getBatch ;
|
||||||
|
stats = [] ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
% Train and validate
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
modelPath = @(ep) fullfile(opts.expDir, sprintf('net-epoch-%d.mat', ep));
|
||||||
|
modelFigPath = fullfile(opts.expDir, 'net-train.pdf') ;
|
||||||
|
|
||||||
|
start = opts.continue * findLastCheckpoint(opts.expDir) ;
|
||||||
|
if start >= 1
|
||||||
|
fprintf('%s: resuming by loading epoch %d\n', mfilename, start) ;
|
||||||
|
[net, stats] = loadState(modelPath(start)) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
if ( opts.computeBNMoment == true )
|
||||||
|
% Validation without using the moving average of BN momemnts
|
||||||
|
for i = 1:numel(net.layers)
|
||||||
|
if ( isa( net.layers(i).block, 'dagnn.BatchNorm') )
|
||||||
|
net.layers(i).block.computeMoment = true;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
|
||||||
|
for epoch=start+1:opts.numEpochs
|
||||||
|
|
||||||
|
% Set the random seed based on the epoch and opts.randomSeed.
|
||||||
|
% This is important for reproducibility, including when training
|
||||||
|
% is restarted from a checkpoint.
|
||||||
|
|
||||||
|
rng(epoch + opts.randomSeed) ;
|
||||||
|
prepareGPUs(opts, epoch == start+1) ;
|
||||||
|
|
||||||
|
% Train for one epoch.
|
||||||
|
state.epoch = epoch ;
|
||||||
|
state.learningRate = opts.learningRate(min(epoch, numel(opts.learningRate))) ;
|
||||||
|
|
||||||
|
%state.train = opts.train(randperm(numel(opts.train))) ; % shuffle
|
||||||
|
%state.val = opts.val(randperm(numel(opts.val))) ;
|
||||||
|
|
||||||
|
% shuffle
|
||||||
|
if( opts.stegoShuffle == 1 )
|
||||||
|
|
||||||
|
N = numel(opts.train);
|
||||||
|
M = numel(opts.val);
|
||||||
|
Lab = max(1, numel(opts.gpus));
|
||||||
|
|
||||||
|
% M and N must be even, and multiple Lab
|
||||||
|
assert( ( rem(N, 2*Lab) == 0 ) & ( rem(M, 2*Lab) == 0 ) );
|
||||||
|
|
||||||
|
% state.train(1:2:N) = opts.train(2*randperm(N/2) - 1);
|
||||||
|
% state.train(2:2:N) = state.train(1:2:N) + 1;
|
||||||
|
%
|
||||||
|
% state.val(1:2:M) = opts.val(2*randperm(M/2) - 1);
|
||||||
|
% state.val(2:2:M) = state.val(1:2:M) + 1;
|
||||||
|
|
||||||
|
seq = opts.train(2*randperm(N/2) - 1);
|
||||||
|
seq = reshape(seq, Lab, N/(2*Lab));
|
||||||
|
state.train = reshape([seq; seq+1], 1, N);
|
||||||
|
|
||||||
|
seq = opts.val(2*randperm(M/2) - 1);
|
||||||
|
seq = reshape(seq, Lab, M/(2*Lab));
|
||||||
|
state.val = reshape([seq; seq+1], 1, M);
|
||||||
|
|
||||||
|
elseif ( opts.stegoShuffle < 0 )
|
||||||
|
% for regression task
|
||||||
|
K = abs( opts.stegoShuffle );
|
||||||
|
|
||||||
|
M = numel(opts.train)/K;
|
||||||
|
seq = K * ( randperm(M) - 1 );
|
||||||
|
seq = [seq + 1; seq + 2; seq + 3; seq + 4; seq + 5; seq + 6];
|
||||||
|
seq = reshape(seq, numel(seq), 1);
|
||||||
|
state.train = opts.train(seq);
|
||||||
|
|
||||||
|
|
||||||
|
N = numel(opts.val)/K;
|
||||||
|
seq = K * ( randperm(N) - 1 ) ;
|
||||||
|
seq = [seq + 1; seq + 2; seq + 3; seq + 4; seq + 5; seq + 6];
|
||||||
|
seq = reshape(seq, numel(seq), 1);
|
||||||
|
state.val = opts.val(seq);
|
||||||
|
|
||||||
|
else
|
||||||
|
state.train = opts.train(randperm(numel(opts.train))) ;
|
||||||
|
state.val = opts.val(randperm(numel(opts.val))) ;
|
||||||
|
end
|
||||||
|
state.imdb = imdb ;
|
||||||
|
|
||||||
|
if numel(opts.gpus) <= 1
|
||||||
|
[stats.train(epoch),prof] = process_epoch(net, state, opts, 'train') ;
|
||||||
|
stats.val(epoch) = process_epoch(net, state, opts, 'val') ;
|
||||||
|
if opts.profile
|
||||||
|
profview(0,prof) ;
|
||||||
|
keyboard ;
|
||||||
|
end
|
||||||
|
else
|
||||||
|
savedNet = net.saveobj() ;
|
||||||
|
spmd
|
||||||
|
net_ = dagnn.DagNN.loadobj(savedNet) ;
|
||||||
|
[stats_.train, prof_] = process_epoch(net_, state, opts, 'train') ;
|
||||||
|
stats_.val = process_epoch(net_, state, opts, 'val') ;
|
||||||
|
if labindex == 1, savedNet_ = net_.saveobj() ; end
|
||||||
|
end
|
||||||
|
net = dagnn.DagNN.loadobj(savedNet_{1}) ;
|
||||||
|
stats__ = accumulateStats(stats_) ;
|
||||||
|
stats.train(epoch) = stats__.train ;
|
||||||
|
stats.val(epoch) = stats__.val ;
|
||||||
|
if opts.profile
|
||||||
|
mpiprofile('viewer', [prof_{:,1}]) ;
|
||||||
|
keyboard ;
|
||||||
|
end
|
||||||
|
clear net_ stats_ stats__ savedNet savedNet_ ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% save
|
||||||
|
if ~evaluateMode
|
||||||
|
saveState(modelPath(epoch), net, stats) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
if opts.plotStatistics
|
||||||
|
switchFigure(1) ; clf ;
|
||||||
|
plots = setdiff(...
|
||||||
|
cat(2,...
|
||||||
|
fieldnames(stats.train)', ...
|
||||||
|
fieldnames(stats.val)'), {'num', 'time'}) ;
|
||||||
|
for p = plots
|
||||||
|
p = char(p) ;
|
||||||
|
values = zeros(0, epoch) ;
|
||||||
|
leg = {} ;
|
||||||
|
for f = {'train', 'val'}
|
||||||
|
f = char(f) ;
|
||||||
|
if isfield(stats.(f), p)
|
||||||
|
tmp = [stats.(f).(p)] ;
|
||||||
|
values(end+1,:) = tmp(1,:)' ;
|
||||||
|
leg{end+1} = f ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
subplot(1,numel(plots),find(strcmp(p,plots))) ;
|
||||||
|
plot(1:epoch, values','o-') ;
|
||||||
|
xlabel('epoch') ;
|
||||||
|
title(p) ;
|
||||||
|
legend(leg{:}) ;
|
||||||
|
grid on ;
|
||||||
|
end
|
||||||
|
drawnow ;
|
||||||
|
print(1, modelFigPath, '-dpdf') ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
if ( opts.computeBNMoment == true )
|
||||||
|
% Revert it back
|
||||||
|
for i = 1:numel(net.layers)
|
||||||
|
if ( isa( net.layers(i).block, 'dagnn.BatchNorm') )
|
||||||
|
net.layers(i).block.computeMoment = false;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function [stats, prof] = process_epoch(net, state, opts, mode)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
% initialize empty momentum
|
||||||
|
if strcmp(mode,'train')
|
||||||
|
state.momentum = num2cell(zeros(1, numel(net.params))) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% move CNN to GPU as needed
|
||||||
|
numGpus = numel(opts.gpus) ;
|
||||||
|
if numGpus >= 1
|
||||||
|
net.move('gpu') ;
|
||||||
|
if strcmp(mode,'train')
|
||||||
|
state.momentum = cellfun(@gpuArray,state.momentum,'UniformOutput',false) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
if numGpus > 1
|
||||||
|
mmap = map_gradients(opts.memoryMapFile, net, numGpus) ;
|
||||||
|
else
|
||||||
|
mmap = [] ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% profile
|
||||||
|
if opts.profile
|
||||||
|
if numGpus <= 1
|
||||||
|
profile clear ;
|
||||||
|
profile on ;
|
||||||
|
else
|
||||||
|
mpiprofile reset ;
|
||||||
|
mpiprofile on ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
subset = state.(mode) ;
|
||||||
|
num = 0 ;
|
||||||
|
stats.num = 0 ; % return something even if subset = []
|
||||||
|
stats.time = 0 ;
|
||||||
|
adjustTime = 0 ;
|
||||||
|
|
||||||
|
start = tic ;
|
||||||
|
for t=1:opts.batchSize:numel(subset)
|
||||||
|
fprintf('%s: epoch %02d: %3d/%3d:', mode, state.epoch, ...
|
||||||
|
fix((t-1)/opts.batchSize)+1, ceil(numel(subset)/opts.batchSize)) ;
|
||||||
|
batchSize = min(opts.batchSize, numel(subset) - t + 1) ;
|
||||||
|
|
||||||
|
for s=1:opts.numSubBatches
|
||||||
|
% get this image batch and prefetch the next
|
||||||
|
batchStart = t + (labindex-1) + (s-1) * numlabs ;
|
||||||
|
batchEnd = min(t+opts.batchSize-1, numel(subset)) ;
|
||||||
|
batch = subset(batchStart : opts.numSubBatches * numlabs : batchEnd) ;
|
||||||
|
num = num + numel(batch) ;
|
||||||
|
if numel(batch) == 0, continue ; end
|
||||||
|
|
||||||
|
inputs = state.getBatch(state.imdb, batch) ;
|
||||||
|
|
||||||
|
if opts.prefetch
|
||||||
|
if s == opts.numSubBatches
|
||||||
|
batchStart = t + (labindex-1) + opts.batchSize ;
|
||||||
|
batchEnd = min(t+2*opts.batchSize-1, numel(subset)) ;
|
||||||
|
else
|
||||||
|
batchStart = batchStart + numlabs ;
|
||||||
|
end
|
||||||
|
nextBatch = subset(batchStart : opts.numSubBatches * numlabs : batchEnd) ;
|
||||||
|
state.getBatch(state.imdb, nextBatch) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
if strcmp(mode, 'train')
|
||||||
|
net.mode = 'normal' ;
|
||||||
|
net.accumulateParamDers = (s ~= 1) ;
|
||||||
|
net.eval(inputs, opts.derOutputs) ;
|
||||||
|
else
|
||||||
|
net.mode = 'test' ;
|
||||||
|
net.eval(inputs) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
% accumulate gradient
|
||||||
|
if strcmp(mode, 'train')
|
||||||
|
if ~isempty(mmap)
|
||||||
|
write_gradients(mmap, net) ;
|
||||||
|
labBarrier() ;
|
||||||
|
end
|
||||||
|
state = accumulate_gradients(state, net, opts, batchSize, mmap) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% get statistics
|
||||||
|
time = toc(start) + adjustTime ;
|
||||||
|
batchTime = time - stats.time ;
|
||||||
|
stats = opts.extractStatsFn(net) ;
|
||||||
|
stats.num = num ;
|
||||||
|
stats.time = time ;
|
||||||
|
currentSpeed = batchSize / batchTime ;
|
||||||
|
averageSpeed = (t + batchSize - 1) / time ;
|
||||||
|
if t == opts.batchSize + 1
|
||||||
|
% compensate for the first iteration, which is an outlier
|
||||||
|
adjustTime = 2*batchTime - time ;
|
||||||
|
stats.time = time + adjustTime ;
|
||||||
|
end
|
||||||
|
|
||||||
|
fprintf(' %.1f (%.1f) Hz', averageSpeed, currentSpeed) ;
|
||||||
|
for f = setdiff(fieldnames(stats)', {'num', 'time'})
|
||||||
|
f = char(f) ;
|
||||||
|
fprintf(' %s:', f) ;
|
||||||
|
fprintf(' %.3f', stats.(f)) ;
|
||||||
|
end
|
||||||
|
fprintf('\n') ;
|
||||||
|
end
|
||||||
|
|
||||||
|
if ~isempty(mmap)
|
||||||
|
unmap_gradients(mmap) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
if opts.profile
|
||||||
|
if numGpus <= 1
|
||||||
|
prof = profile('info') ;
|
||||||
|
profile off ;
|
||||||
|
else
|
||||||
|
prof = mpiprofile('info');
|
||||||
|
mpiprofile off ;
|
||||||
|
end
|
||||||
|
else
|
||||||
|
prof = [] ;
|
||||||
|
end
|
||||||
|
|
||||||
|
net.reset() ;
|
||||||
|
net.move('cpu') ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function state = accumulate_gradients(state, net, opts, batchSize, mmap)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
numGpus = numel(opts.gpus) ;
|
||||||
|
otherGpus = setdiff(1:numGpus, labindex) ;
|
||||||
|
|
||||||
|
for p=1:numel(net.params)
|
||||||
|
|
||||||
|
% accumualte gradients from multiple labs (GPUs) if needed
|
||||||
|
if numGpus > 1
|
||||||
|
tag = net.params(p).name ;
|
||||||
|
for g = otherGpus
|
||||||
|
tmp = gpuArray(mmap.Data(g).(tag)) ;
|
||||||
|
net.params(p).der = net.params(p).der + tmp ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
switch net.params(p).trainMethod
|
||||||
|
|
||||||
|
case 'average' % mainly for batch normalization
|
||||||
|
thisLR = net.params(p).learningRate ;
|
||||||
|
net.params(p).value = ...
|
||||||
|
(1 - thisLR) * net.params(p).value + ...
|
||||||
|
(thisLR/batchSize/net.params(p).fanout) * net.params(p).der ;
|
||||||
|
|
||||||
|
case 'gradient'
|
||||||
|
thisDecay = opts.weightDecay * net.params(p).weightDecay ;
|
||||||
|
thisLR = state.learningRate * net.params(p).learningRate ;
|
||||||
|
state.momentum{p} = opts.momentum * state.momentum{p} ...
|
||||||
|
- thisDecay * net.params(p).value ...
|
||||||
|
- (1 / batchSize) * net.params(p).der ;
|
||||||
|
net.params(p).value = net.params(p).value + thisLR * state.momentum{p} ;
|
||||||
|
|
||||||
|
case 'otherwise'
|
||||||
|
error('Unknown training method ''%s'' for parameter ''%s''.', ...
|
||||||
|
net.params(p).trainMethod, ...
|
||||||
|
net.params(p).name) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function mmap = map_gradients(fname, net, numGpus)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
format = {} ;
|
||||||
|
for i=1:numel(net.params)
|
||||||
|
format(end+1,1:3) = {'single', size(net.params(i).value), net.params(i).name} ;
|
||||||
|
end
|
||||||
|
format(end+1,1:3) = {'double', [3 1], 'errors'} ;
|
||||||
|
if ~exist(fname) && (labindex == 1)
|
||||||
|
f = fopen(fname,'wb') ;
|
||||||
|
for g=1:numGpus
|
||||||
|
for i=1:size(format,1)
|
||||||
|
fwrite(f,zeros(format{i,2},format{i,1}),format{i,1}) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
fclose(f) ;
|
||||||
|
end
|
||||||
|
labBarrier() ;
|
||||||
|
mmap = memmapfile(fname, ...
|
||||||
|
'Format', format, ...
|
||||||
|
'Repeat', numGpus, ...
|
||||||
|
'Writable', true) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function write_gradients(mmap, net)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
for i=1:numel(net.params)
|
||||||
|
mmap.Data(labindex).(net.params(i).name) = gather(net.params(i).der) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function unmap_gradients(mmap)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function stats = accumulateStats(stats_)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
|
||||||
|
for s = {'train', 'val'}
|
||||||
|
s = char(s) ;
|
||||||
|
total = 0 ;
|
||||||
|
|
||||||
|
% initialize stats stucture with same fields and same order as
|
||||||
|
% stats_{1}
|
||||||
|
stats__ = stats_{1} ;
|
||||||
|
names = fieldnames(stats__.(s))' ;
|
||||||
|
values = zeros(1, numel(names)) ;
|
||||||
|
fields = cat(1, names, num2cell(values)) ;
|
||||||
|
stats.(s) = struct(fields{:}) ;
|
||||||
|
|
||||||
|
for g = 1:numel(stats_)
|
||||||
|
stats__ = stats_{g} ;
|
||||||
|
num__ = stats__.(s).num ;
|
||||||
|
total = total + num__ ;
|
||||||
|
|
||||||
|
for f = setdiff(fieldnames(stats__.(s))', 'num')
|
||||||
|
f = char(f) ;
|
||||||
|
stats.(s).(f) = stats.(s).(f) + stats__.(s).(f) * num__ ;
|
||||||
|
|
||||||
|
if g == numel(stats_)
|
||||||
|
stats.(s).(f) = stats.(s).(f) / total ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
stats.(s).num = total ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function stats = extractStats(net)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
sel = find(cellfun(@(x) isa(x,'dagnn.Loss'), {net.layers.block})) ;
|
||||||
|
stats = struct() ;
|
||||||
|
for i = 1:numel(sel)
|
||||||
|
stats.(net.layers(sel(i)).outputs{1}) = net.layers(sel(i)).block.average ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function saveState(fileName, net, stats)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
net_ = net ;
|
||||||
|
net = net_.saveobj() ;
|
||||||
|
save(fileName, 'net', 'stats') ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function [net, stats] = loadState(fileName)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
load(fileName, 'net', 'stats') ;
|
||||||
|
net = dagnn.DagNN.loadobj(net) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function epoch = findLastCheckpoint(modelDir)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
list = dir(fullfile(modelDir, 'net-epoch-*.mat')) ;
|
||||||
|
tokens = regexp({list.name}, 'net-epoch-([\d]+).mat', 'tokens') ;
|
||||||
|
epoch = cellfun(@(x) sscanf(x{1}{1}, '%d'), tokens) ;
|
||||||
|
epoch = max([epoch 0]) ;
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function switchFigure(n)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
if get(0,'CurrentFigure') ~= n
|
||||||
|
try
|
||||||
|
set(0,'CurrentFigure',n) ;
|
||||||
|
catch
|
||||||
|
figure(n) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
function prepareGPUs(opts, cold)
|
||||||
|
% -------------------------------------------------------------------------
|
||||||
|
numGpus = numel(opts.gpus) ;
|
||||||
|
if numGpus > 1
|
||||||
|
% check parallel pool integrity as it could have timed out
|
||||||
|
pool = gcp('nocreate') ;
|
||||||
|
if ~isempty(pool) && pool.NumWorkers ~= numGpus
|
||||||
|
delete(pool) ;
|
||||||
|
end
|
||||||
|
pool = gcp('nocreate') ;
|
||||||
|
if isempty(pool)
|
||||||
|
parpool('local', numGpus) ;
|
||||||
|
cold = true ;
|
||||||
|
end
|
||||||
|
if exist(opts.memoryMapFile)
|
||||||
|
delete(opts.memoryMapFile) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
end
|
||||||
|
if numGpus >= 1 && cold
|
||||||
|
fprintf('%s: resetting GPU\n', mfilename)
|
||||||
|
if numGpus == 1
|
||||||
|
gpuDevice(opts.gpus)
|
||||||
|
else
|
||||||
|
spmd, gpuDevice(opts.gpus(labindex)), end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
%end
|
||||||
|
|
@ -0,0 +1,12 @@
|
||||||
|
classdef Abs < dagnn.ElementWise
|
||||||
|
methods
|
||||||
|
function outputs = forward(obj, inputs, params)
|
||||||
|
outputs{1} = vl_nnabs(inputs{1}) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
function [derInputs, derParams] = backward(obj, inputs, params, derOutputs)
|
||||||
|
derInputs{1} = vl_nnabs(inputs{1}, derOutputs{1}) ;
|
||||||
|
derParams = {} ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,60 @@
|
||||||
|
classdef BatchNorm < dagnn.ElementWise
|
||||||
|
properties
|
||||||
|
numChannels
|
||||||
|
epsilon = 1e-4
|
||||||
|
computeMoment = false;
|
||||||
|
end
|
||||||
|
|
||||||
|
properties (Transient)
|
||||||
|
moments
|
||||||
|
end
|
||||||
|
|
||||||
|
methods
|
||||||
|
function outputs = forward(obj, inputs, params)
|
||||||
|
if strcmp(obj.net.mode, 'test')
|
||||||
|
if( obj.computeMoment )
|
||||||
|
[outputs{1}, obj.moments] = vl_nnbnorm(inputs{1}, params{1}, ...
|
||||||
|
params{2}, 'epsilon', obj.epsilon) ;
|
||||||
|
else
|
||||||
|
outputs{1} = vl_nnbnorm(inputs{1}, params{1}, params{2}, ...
|
||||||
|
'moments', params{3}, ...
|
||||||
|
'epsilon', obj.epsilon) ;
|
||||||
|
obj.moments = [];
|
||||||
|
end
|
||||||
|
|
||||||
|
else
|
||||||
|
[outputs{1}, obj.moments] = vl_nnbnorm(inputs{1}, params{1}, ...
|
||||||
|
params{2}, 'epsilon', obj.epsilon) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
function [derInputs, derParams] = backward(obj, inputs, params, derOutputs)
|
||||||
|
[derInputs{1}, derParams{1}, derParams{2}, derParams{3}] = ...
|
||||||
|
vl_nnbnorm(inputs{1}, params{1}, params{2}, derOutputs{1}, ...
|
||||||
|
'epsilon', obj.epsilon) ;
|
||||||
|
obj.moments = [];
|
||||||
|
% multiply the moments update by the number of images in the batch
|
||||||
|
% this is required to make the update additive for subbatches
|
||||||
|
% and will eventually be normalized away
|
||||||
|
derParams{3} = derParams{3} * size(inputs{1},4) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
% ---------------------------------------------------------------------
|
||||||
|
function obj = BatchNorm(varargin)
|
||||||
|
obj.load(varargin{:}) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
function params = initParams(obj)
|
||||||
|
params{1} = ones(obj.numChannels,1,'single') ;
|
||||||
|
params{2} = zeros(obj.numChannels,1,'single') ;
|
||||||
|
params{3} = zeros(obj.numChannels,2,'single') ;
|
||||||
|
end
|
||||||
|
|
||||||
|
function attach(obj, net, index)
|
||||||
|
attach@dagnn.ElementWise(obj, net, index) ;
|
||||||
|
p = net.getParamIndex(net.layers(index).params{3}) ;
|
||||||
|
net.params(p).trainMethod = 'average' ;
|
||||||
|
net.params(p).learningRate = 0.01 ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,36 @@
|
||||||
|
classdef PhaseSplit < dagnn.Filter
|
||||||
|
% Construct PhaseSplit in a way similar to Pooling, mightbe we could
|
||||||
|
% do it in a better and clean way.
|
||||||
|
properties
|
||||||
|
poolSize = [1 1]
|
||||||
|
end
|
||||||
|
|
||||||
|
methods
|
||||||
|
function outputs = forward(self, inputs, params)
|
||||||
|
outputs{1} = vl_nnphasesplit( inputs{1} ) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
function [derInputs, derParams] = backward(self, inputs, params, derOutputs)
|
||||||
|
derInputs{1} = vl_nnphasesplit( inputs{1}, derOutputs{1} ) ;
|
||||||
|
derParams = {} ;
|
||||||
|
end
|
||||||
|
|
||||||
|
function kernelSize = getKernelSize(obj)
|
||||||
|
kernelSize = obj.poolSize ;
|
||||||
|
end
|
||||||
|
|
||||||
|
function outputSizes = getOutputSizes(obj, inputSizes)
|
||||||
|
%outputSizes = getOutputSizes@dagnn.Filter(obj, inputSizes) ;
|
||||||
|
outputSizes{1}(1) = inputSizes{1}(1)/8 ;
|
||||||
|
outputSizes{1}(2) = inputSizes{1}(2)/8;
|
||||||
|
outputSizes{1}(3) = inputSizes{1}(3)*64;
|
||||||
|
outputSizes{1}(4) = inputSizes{1}(4);
|
||||||
|
end
|
||||||
|
|
||||||
|
function obj = PhaseSplit(varargin)
|
||||||
|
%obj.load(varargin) ;
|
||||||
|
obj.pad = [0 0 0 0];
|
||||||
|
obj.stride = [8 8];
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,12 @@
|
||||||
|
classdef TanH < dagnn.ElementWise
|
||||||
|
methods
|
||||||
|
function outputs = forward(obj, inputs, params)
|
||||||
|
outputs{1} = vl_nntanh(inputs{1}) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
function [derInputs, derParams] = backward(obj, inputs, params, derOutputs)
|
||||||
|
derInputs{1} = vl_nntanh(inputs{1}, derOutputs{1}) ;
|
||||||
|
derParams = {} ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,236 @@
|
||||||
|
function obj = fromSimpleNN(net, varargin)
|
||||||
|
% FROMSIMPLENN Initialize a DagNN object from a SimpleNN network
|
||||||
|
% FROMSIMPLENN(NET) initializes the DagNN object from the
|
||||||
|
% specified CNN using the SimpleNN format.
|
||||||
|
%
|
||||||
|
% SimpleNN objects are linear chains of computational layers. These
|
||||||
|
% layers exchange information through variables and parameters that
|
||||||
|
% are not explicitly named. Hence, FROMSIMPLENN() uses a number of
|
||||||
|
% rules to assign such names automatically:
|
||||||
|
%
|
||||||
|
% * From the input to the output of the CNN, variables are called
|
||||||
|
% `x0` (input of the first layer), `x1`, `x2`, .... In this
|
||||||
|
% manner `xi` is the output of the i-th layer.
|
||||||
|
%
|
||||||
|
% * Any loss layer requires two inputs, the second being a label.
|
||||||
|
% These are called `label` (for the first such layers), and then
|
||||||
|
% `label2`, `label3`,... for any other similar layer.
|
||||||
|
%
|
||||||
|
% Additionally, given the option `CanonicalNames` the function can
|
||||||
|
% change the names of some variables to make them more convenient to
|
||||||
|
% use. With this option turned on:
|
||||||
|
%
|
||||||
|
% * The network input is called `input` instead of `x0`.
|
||||||
|
%
|
||||||
|
% * The output of each SoftMax layer is called `prob` (or `prob2`,
|
||||||
|
% ...).
|
||||||
|
%
|
||||||
|
% * The output of each Loss layer is called `objective` (or `
|
||||||
|
% objective2`, ...).
|
||||||
|
%
|
||||||
|
% * The input of each SoftMax or Loss layer of type *softmax log
|
||||||
|
% loss* is called `prediction` (or `prediction2`, ...). If a Loss
|
||||||
|
% layer immediately follows a SoftMax layer, then the rule above
|
||||||
|
% takes precendence and the input name is not changed.
|
||||||
|
%
|
||||||
|
% FROMSIMPLENN(___, 'OPT', VAL, ...) accepts the following options:
|
||||||
|
%
|
||||||
|
% `CanonicalNames`:: false
|
||||||
|
% If `true` use the rules above to assign more meaningful
|
||||||
|
% names to some of the variables.
|
||||||
|
|
||||||
|
% Copyright (C) 2015 Karel Lenc and Andrea Vedaldi.
|
||||||
|
% All rights reserved.
|
||||||
|
%
|
||||||
|
% This file is part of the VLFeat library and is made available under
|
||||||
|
% the terms of the BSD license (see the COPYING file).
|
||||||
|
|
||||||
|
opts.canonicalNames = false ;
|
||||||
|
opts = vl_argparse(opts, varargin) ;
|
||||||
|
|
||||||
|
import dagnn.*
|
||||||
|
|
||||||
|
obj = DagNN() ;
|
||||||
|
net = vl_simplenn_move(net, 'cpu') ;
|
||||||
|
net = vl_simplenn_tidy(net) ;
|
||||||
|
|
||||||
|
% copy meta-information as is
|
||||||
|
obj.meta = net.meta ;
|
||||||
|
|
||||||
|
for l = 1:numel(net.layers)
|
||||||
|
inputs = {sprintf('x%d',l-1)} ;
|
||||||
|
outputs = {sprintf('x%d',l)} ;
|
||||||
|
|
||||||
|
params = struct(...
|
||||||
|
'name', {}, ...
|
||||||
|
'value', {}, ...
|
||||||
|
'learningRate', [], ...
|
||||||
|
'weightDecay', []) ;
|
||||||
|
if isfield(net.layers{l}, 'name')
|
||||||
|
name = net.layers{l}.name ;
|
||||||
|
else
|
||||||
|
name = sprintf('layer%d',l) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
switch net.layers{l}.type
|
||||||
|
case {'conv', 'convt'}
|
||||||
|
sz = size(net.layers{l}.weights{1}) ;
|
||||||
|
hasBias = ~isempty(net.layers{l}.weights{2}) ;
|
||||||
|
params(1).name = sprintf('%sf',name) ;
|
||||||
|
params(1).value = net.layers{l}.weights{1} ;
|
||||||
|
if hasBias
|
||||||
|
params(2).name = sprintf('%sb',name) ;
|
||||||
|
params(2).value = net.layers{l}.weights{2} ;
|
||||||
|
end
|
||||||
|
if isfield(net.layers{l},'learningRate')
|
||||||
|
params(1).learningRate = net.layers{l}.learningRate(1) ;
|
||||||
|
if hasBias
|
||||||
|
params(2).learningRate = net.layers{l}.learningRate(2) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
if isfield(net.layers{l},'weightDecay')
|
||||||
|
params(1).weightDecay = net.layers{l}.weightDecay(1) ;
|
||||||
|
if hasBias
|
||||||
|
params(2).weightDecay = net.layers{l}.weightDecay(2) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
switch net.layers{l}.type
|
||||||
|
case 'conv'
|
||||||
|
block = Conv() ;
|
||||||
|
block.size = sz ;
|
||||||
|
block.pad = net.layers{l}.pad ;
|
||||||
|
block.stride = net.layers{l}.stride ;
|
||||||
|
case 'convt'
|
||||||
|
block = ConvTranspose() ;
|
||||||
|
block.size = sz ;
|
||||||
|
block.upsample = net.layers{l}.upsample ;
|
||||||
|
block.crop = net.layers{l}.crop ;
|
||||||
|
block.numGroups = net.layers{l}.numGroups ;
|
||||||
|
end
|
||||||
|
block.hasBias = hasBias ;
|
||||||
|
block.opts = net.layers{l}.opts ;
|
||||||
|
|
||||||
|
case 'pool'
|
||||||
|
block = Pooling() ;
|
||||||
|
block.method = net.layers{l}.method ;
|
||||||
|
block.poolSize = net.layers{l}.pool ;
|
||||||
|
block.pad = net.layers{l}.pad ;
|
||||||
|
block.stride = net.layers{l}.stride ;
|
||||||
|
block.opts = net.layers{l}.opts ;
|
||||||
|
|
||||||
|
case 'phasesplit'
|
||||||
|
block = PhaseSplit();
|
||||||
|
|
||||||
|
case {'normalize', 'lrn'}
|
||||||
|
block = LRN() ;
|
||||||
|
block.param = net.layers{l}.param ;
|
||||||
|
|
||||||
|
case {'dropout'}
|
||||||
|
block = DropOut() ;
|
||||||
|
block.rate = net.layers{l}.rate ;
|
||||||
|
|
||||||
|
case {'relu'}
|
||||||
|
block = ReLU() ;
|
||||||
|
block.leak = net.layers{l}.leak ;
|
||||||
|
|
||||||
|
case {'sigmoid'}
|
||||||
|
block = Sigmoid() ;
|
||||||
|
|
||||||
|
case {'abs'}
|
||||||
|
block = Abs();
|
||||||
|
|
||||||
|
case {'tanh'}
|
||||||
|
block = TanH();
|
||||||
|
|
||||||
|
case {'tlu'}
|
||||||
|
block = TLU();
|
||||||
|
|
||||||
|
case {'softmax'}
|
||||||
|
block = SoftMax() ;
|
||||||
|
|
||||||
|
case {'softmaxloss'}
|
||||||
|
block = Loss('loss', 'softmaxlog') ;
|
||||||
|
% The loss has two inputs
|
||||||
|
inputs{2} = getNewVarName(obj, 'label') ;
|
||||||
|
|
||||||
|
case {'l2'}
|
||||||
|
block = Loss('loss', 'l2') ;
|
||||||
|
% The loss has two inputs
|
||||||
|
inputs{2} = getNewVarName(obj, 'label') ;
|
||||||
|
|
||||||
|
case {'bnorm'}
|
||||||
|
block = BatchNorm() ;
|
||||||
|
params(1).name = sprintf('%sm',name) ;
|
||||||
|
params(1).value = net.layers{l}.weights{1} ;
|
||||||
|
params(2).name = sprintf('%sb',name) ;
|
||||||
|
params(2).value = net.layers{l}.weights{2} ;
|
||||||
|
params(3).name = sprintf('%sx',name) ;
|
||||||
|
params(3).value = net.layers{l}.weights{3} ;
|
||||||
|
if isfield(net.layers{l},'learningRate')
|
||||||
|
params(1).learningRate = net.layers{l}.learningRate(1) ;
|
||||||
|
params(2).learningRate = net.layers{l}.learningRate(2) ;
|
||||||
|
params(3).learningRate = net.layers{l}.learningRate(3) ;
|
||||||
|
end
|
||||||
|
if isfield(net.layers{l},'weightDecay')
|
||||||
|
params(1).weightDecay = net.layers{l}.weightDecay(1) ;
|
||||||
|
params(2).weightDecay = net.layers{l}.weightDecay(2) ;
|
||||||
|
params(3).weightDecay = 0 ;
|
||||||
|
end
|
||||||
|
|
||||||
|
otherwise
|
||||||
|
error([net.layers{l}.type ' is unsupported']) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
obj.addLayer(...
|
||||||
|
name, ...
|
||||||
|
block, ...
|
||||||
|
inputs, ...
|
||||||
|
outputs, ...
|
||||||
|
{params.name}) ;
|
||||||
|
|
||||||
|
for p = 1:numel(params)
|
||||||
|
pindex = obj.getParamIndex(params(p).name) ;
|
||||||
|
if ~isempty(params(p).value)
|
||||||
|
obj.params(pindex).value = params(p).value ;
|
||||||
|
end
|
||||||
|
if ~isempty(params(p).learningRate)
|
||||||
|
obj.params(pindex).learningRate = params(p).learningRate ;
|
||||||
|
end
|
||||||
|
if ~isempty(params(p).weightDecay)
|
||||||
|
obj.params(pindex).weightDecay = params(p).weightDecay ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
% --------------------------------------------------------------------
|
||||||
|
% Rename variables to canonical names
|
||||||
|
% --------------------------------------------------------------------
|
||||||
|
|
||||||
|
if opts.canonicalNames
|
||||||
|
for l = 1:numel(obj.layers)
|
||||||
|
if l == 1
|
||||||
|
obj.renameVar(obj.layers(l).inputs{1}, 'input') ;
|
||||||
|
end
|
||||||
|
if isa(obj.layers(l).block, 'dagnn.SoftMax')
|
||||||
|
obj.renameVar(obj.layers(l).outputs{1}, getNewVarName(obj, 'prob')) ;
|
||||||
|
obj.renameVar(obj.layers(l).inputs{1}, getNewVarName(obj, 'prediction')) ;
|
||||||
|
end
|
||||||
|
if isa(obj.layers(l).block, 'dagnn.Loss') %|| isa(obj.layers(l).block, 'dagnn.L2Dist')
|
||||||
|
obj.renameVar(obj.layers(l).outputs{1}, 'objective') ;
|
||||||
|
if isempty(regexp(obj.layers(l).inputs{1}, '^prob.*'))
|
||||||
|
obj.renameVar(obj.layers(l).inputs{1}, ...
|
||||||
|
getNewVarName(obj, 'prediction')) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
% --------------------------------------------------------------------
|
||||||
|
function name = getNewVarName(obj, prefix)
|
||||||
|
% --------------------------------------------------------------------
|
||||||
|
t = 0 ;
|
||||||
|
name = prefix ;
|
||||||
|
while any(strcmp(name, {obj.vars.name}))
|
||||||
|
t = t + 1 ;
|
||||||
|
name = sprintf('%s%d', prefix, t) ;
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,19 @@
|
||||||
|
function out = vl_nnabs(x,dzdy)
|
||||||
|
%VL_NNABS CNN ABS unit.
|
||||||
|
% Y = VL_NNABS(X) computes the absolute value of the data X. X can
|
||||||
|
% have an arbitrary size. The abs is defined as follows:
|
||||||
|
%
|
||||||
|
% ABS(X) = |X|.
|
||||||
|
%
|
||||||
|
% DZDX = VL_NNABS(X, DZDY) computes the derivative of the
|
||||||
|
% block projected onto DZDY. DZDX and DZDY have the same
|
||||||
|
% dimensions as X and Y respectively.
|
||||||
|
|
||||||
|
% Note, MATLAB built-in function ABS() and SIGN() are used because their
|
||||||
|
% support for gpuArray
|
||||||
|
|
||||||
|
if nargin <= 1 || isempty(dzdy)
|
||||||
|
out = abs( x ) ;
|
||||||
|
else
|
||||||
|
out = dzdy .* sign( x );
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,58 @@
|
||||||
|
function y = vl_nnphasesplit( x, dzdy )
|
||||||
|
%VL_NNPHASESPLIT CNN phase split the feature plane into 8x8 = 64 DCT mode.
|
||||||
|
% Y = VL_NNCROP(X, STRIDE) phase split the input X into 64 DCT phase mode.
|
||||||
|
%
|
||||||
|
% DZDX = VL_NNCROP(X, DZDY) computes the derivative DZDX of the
|
||||||
|
% function projected on the output derivative DZDY. DZDX has the same
|
||||||
|
% dimension as X and DZDY the same dimension as Y.
|
||||||
|
%
|
||||||
|
|
||||||
|
% dimension must be divided by 8
|
||||||
|
assert( rem(size(x,1), 8 ) == 0 & rem( size(x,2), 8) == 0 );
|
||||||
|
|
||||||
|
% Initialize some parameters
|
||||||
|
inputSize = [size(x,1) size(x,2) size(x,3) size(x,4)] ;
|
||||||
|
outputSize = [size(x,1)/8, size(x,2)/8, 64*size(x,3), size(x,4)];
|
||||||
|
|
||||||
|
% zig zag order
|
||||||
|
zzag = zeros(64, 4);
|
||||||
|
idx = 1;
|
||||||
|
startCh = 1;
|
||||||
|
for i = 0:7
|
||||||
|
for j = 0:7
|
||||||
|
stopCh = startCh + inputSize(3);
|
||||||
|
zzag(idx, :) = [ i, j, startCh, stopCh - 1 ];
|
||||||
|
idx = idx + 1;
|
||||||
|
startCh = stopCh;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
% sampling array
|
||||||
|
sy = 1:8:inputSize(1);
|
||||||
|
sx = 1:8:inputSize(2);
|
||||||
|
|
||||||
|
if nargin <= 1 || isempty(dzdy)
|
||||||
|
% forward function
|
||||||
|
if isa( x, 'gpuArray' )
|
||||||
|
y = gpuArray.zeros(outputSize, classUnderlying(x)) ;
|
||||||
|
else
|
||||||
|
y = zeros(outputSize, 'like', x ) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
for i = 1:64
|
||||||
|
y(:,:,zzag(i,3):zzag(i,4),:) = x(sy + zzag(i,1), sx + zzag(i,2), :, : );
|
||||||
|
end
|
||||||
|
|
||||||
|
else
|
||||||
|
% backward function
|
||||||
|
if isa(dzdy, 'gpuArray')
|
||||||
|
y = gpuArray.zeros(inputSize, classUnderlying(dzdy)) ;
|
||||||
|
else
|
||||||
|
y = zeros(inputSize, 'like', x) ;
|
||||||
|
end
|
||||||
|
|
||||||
|
for i = 1:64
|
||||||
|
y(sy + zzag(i,1), sx + zzag(i,2), :, : ) = dzdy(:,:,zzag(i,3):zzag(i,4),:);
|
||||||
|
end
|
||||||
|
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,21 @@
|
||||||
|
function out = vl_nntanh(x,dzdy)
|
||||||
|
%VL_NNTANH CNN TanH hyperbolic non-linearity
|
||||||
|
% Y = VL_NNTANH(X) computes the hyperbolic tangent non-linearity of the
|
||||||
|
% data X. X can have an arbitrary size. The tanh is defined as follows:
|
||||||
|
%
|
||||||
|
% TANH(X) = (EXP(2X) - 1 )/( EXP(2x) + 1 ).
|
||||||
|
%
|
||||||
|
% DZDX = VL_NNTANH(X, DZDY) computes the derivative of the
|
||||||
|
% block projected onto DZDY. DZDX and DZDY have the same
|
||||||
|
% dimensions as X and Y respectively.
|
||||||
|
%
|
||||||
|
% NOTE: Matlab build-in function TANH() is used since it has extended
|
||||||
|
% support for gpuArray
|
||||||
|
|
||||||
|
y = tanh( x );
|
||||||
|
|
||||||
|
if nargin <= 1 || isempty(dzdy)
|
||||||
|
out = y;
|
||||||
|
else
|
||||||
|
out = dzdy .* ( 1 - y.*y );
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,36 @@
|
||||||
|
classdef nnabs < nntest
|
||||||
|
properties
|
||||||
|
x
|
||||||
|
delta
|
||||||
|
end
|
||||||
|
|
||||||
|
methods (TestClassSetup)
|
||||||
|
function data(test,device)
|
||||||
|
% make sure that all elements in x are differentiable. in this way,
|
||||||
|
% we can compute numerical derivatives reliably by adding a delta < .5.
|
||||||
|
delta = 0.01;
|
||||||
|
test.range = 10 ;
|
||||||
|
x = test.randn(15,14,3,2) ;
|
||||||
|
|
||||||
|
ind = find(( x < 0 )&( x > -2*delta));
|
||||||
|
if (~isempty(ind))
|
||||||
|
x(ind) = -2 + rand([1, length(ind)], 'like', x);
|
||||||
|
end
|
||||||
|
|
||||||
|
test.x = x ;
|
||||||
|
test.delta = delta;
|
||||||
|
|
||||||
|
if strcmp(device,'gpu'), test.x = gpuArray(test.x) ; end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
methods (Test)
|
||||||
|
function basic(test)
|
||||||
|
x = test.x ;
|
||||||
|
y = vl_nnabs(x) ;
|
||||||
|
dzdy = test.randn(size(y)) ;
|
||||||
|
dzdx = vl_nnabs(x,dzdy) ;
|
||||||
|
test.der(@(x) vl_nnabs(x), x, dzdy, dzdx, 1e-2) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,23 @@
|
||||||
|
classdef nnphasesplit < nntest
|
||||||
|
properties
|
||||||
|
x
|
||||||
|
end
|
||||||
|
|
||||||
|
methods (TestClassSetup)
|
||||||
|
function data(test,device)
|
||||||
|
test.range = 10 ;
|
||||||
|
test.x = test.randn(32,32,4,2) ;
|
||||||
|
if strcmp(device,'gpu'), test.x = gpuArray(test.x) ; end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
methods (Test)
|
||||||
|
function basic(test)
|
||||||
|
x = test.x ;
|
||||||
|
y = vl_nnphasesplit(x) ;
|
||||||
|
dzdy = test.randn(size(y)) ;
|
||||||
|
dzdx = vl_nnphasesplit(x,dzdy) ;
|
||||||
|
test.der(@(x) vl_nnphasesplit(x), x, dzdy, dzdx, 1e-3) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
@ -0,0 +1,24 @@
|
||||||
|
classdef nntanh < nntest
|
||||||
|
properties
|
||||||
|
x
|
||||||
|
delta
|
||||||
|
end
|
||||||
|
|
||||||
|
methods (TestClassSetup)
|
||||||
|
function data(test,device)
|
||||||
|
test.range = 10 ;
|
||||||
|
test.x = test.randn(15,14,3,2) ;
|
||||||
|
if strcmp(device,'gpu'), test.x = gpuArray(test.x) ; end
|
||||||
|
end
|
||||||
|
end
|
||||||
|
|
||||||
|
methods (Test)
|
||||||
|
function basic(test)
|
||||||
|
x = test.x ;
|
||||||
|
y = vl_nntanh(x) ;
|
||||||
|
dzdy = test.randn(size(y)) ;
|
||||||
|
dzdx = vl_nntanh(x,dzdy) ;
|
||||||
|
test.der(@(x) vl_nntanh(x), x, dzdy, dzdx, 1e-2) ;
|
||||||
|
end
|
||||||
|
end
|
||||||
|
end
|
||||||
Binary file not shown.
135
README.md
135
README.md
|
|
@ -1,3 +1,134 @@
|
||||||
# Steganalysis
|
# Steganography
|
||||||
|
|
||||||
Python 实现LSB算法进行信息隐藏 包含空域与变换域 JPEG信息隐藏算法 对PDF文件进行信息隐藏 基于卷积神经网络的隐写分析 Matlab SRM、SCA隐写分析
|
# 空域编码图像
|
||||||
|
|
||||||
|
* 空域编码是指在图像空间域进行编码,也就是直接针对图像像素进行编码
|
||||||
|
* 对像素进行编码,如 LSB 算法,主要有下面两种方式
|
||||||
|
* 光栅格式
|
||||||
|
* 调色板格式 GIF(graphics interchange format)
|
||||||
|
* 一个图像编码标准往往包括多类编码方法,一个图像仅仅是其一类方法的实例。例如,常见的 BMP(Bitmap)、 TIFF(
|
||||||
|
Tagged Image File Format)、 PNG(Portable Network
|
||||||
|
Graphics)均支持光栅格式与调色板格式编码,对这两种格式
|
||||||
|
编码分别又支持多种具体编码方法
|
||||||
|
|
||||||
|
## LSB 隐写算法
|
||||||
|
|
||||||
|
* LSB 隐写是最基础、最简单的隐写方法,具有容量大、嵌入速度快、对载体图像质量影响小的特点
|
||||||
|
* LSB 的大意就是最低比特位隐写。我们将深度为 8 的 BMP 图像,分为 8 个二值平面(位平面),我们将待嵌入的信息(info)直接写到最低的位平面上。换句话说,如果秘密信息与最低比特位相同,则不改动;如果秘密信息与最低比特位不同,则使用秘密信息值代替最低比特位
|
||||||
|
|
||||||
|
[代码实现](/LSB)
|
||||||
|
|
||||||
|

|
||||||
|
嵌入信息前的载体图片
|
||||||
|
|
||||||
|

|
||||||
|
嵌入信息后的载体图片
|
||||||
|
|
||||||
|
# 变换域编码图像
|
||||||
|
|
||||||
|
## JPEG
|
||||||
|
|
||||||
|
* Joint Photographic Experts Group(联合图像专家小组)的缩写
|
||||||
|
* JPEG 编码
|
||||||
|

|
||||||
|
|
||||||
|
### JSteg 隐写
|
||||||
|
|
||||||
|
* JSteg 的算法的主要思想是将秘密消息嵌入在量化后的 DCT 系数的最低比特位上,但对原始值为 0、+1、-1 的 DCT 系数不进行嵌入,提取秘密消息时,只需将载密图像中不等于 0、l 的量化 DCT 系数的 LSB 取出即可
|
||||||
|
* JSteg 算法步骤
|
||||||
|
|
||||||
|
1. 选择载体图像,并且将载体图像划分为连续的 8×8 的子块。
|
||||||
|
2. 对每个子块使用离散余弦变换之后,用相应的质量因数的量化表量化,得到对应的 8×8 量化 DCT 子块。
|
||||||
|
3. 将需要隐藏的信息编码为二进制数据流,对 DCT 子块系数进行 Z 字形扫描,并且使用秘密信息的二进制流替换非 0 和非 1 的 DCT 系数的最低比特位。
|
||||||
|
4. 进行熵编码等,产生 JPEG 隐密图像。
|
||||||
|
|
||||||
|
* JSteg 的具体嵌入过程
|
||||||
|
|
||||||
|
1. 部分解码 JPEG 图像,得到二进制存储的 AC 系数,判断该 AC 系数是否等于正负 1 或 0,若等于则跳过该 AC 系数,否则,执行下一步
|
||||||
|
2. 判断二进制存储的 AC 系数的 LSB 是否与要嵌入的秘密信息比特相同,若相同,则不对其进行修改,否则执行下一步
|
||||||
|
3. 用秘密信息比特替换二进制存储的 AC 系数的 LSB,将修改后的 AC 系数重新编码得到隐秘 JPEG 图像
|
||||||
|
|
||||||
|
* JSteg 不使用 0、1 的原因
|
||||||
|
|
||||||
|
1. DCT 系数中“0”的比例最大(一般可达到 60% 以上,取决于图像质量和压缩因子),压缩编码是利用大量出现连零实现的,如果改变 DCT 系数中“0”的话,不能很好的实现压缩
|
||||||
|
2. DCT 系数中的“1”若变成“0”,由于接受端无法区分未使用的“0”和嵌入消息后得到的“0”,从而无法实现秘密信息的提取
|
||||||
|
|
||||||
|
[代码实现](Jsteg.py)
|
||||||
|
|
||||||
|
### F3 隐写
|
||||||
|
|
||||||
|
* 为了改善大量 DCT 系数不隐藏信息这一状况,人们提出了 F3 隐写
|
||||||
|
* F3 对原始值为 +1 和-1 的 DCT 系数,进行了利用。F3 隐写的规则如下
|
||||||
|
|
||||||
|
1. 每个非 0 的 DCT 数据用于隐藏 1 比特秘密信息,为 0 的 DCT 系数不负载秘密信息
|
||||||
|
2. 如果秘密信息与 DCT 的 LSB 相同,便不作改动;如果不同,将 DCT 系数的绝对值减小 1,符号不变
|
||||||
|
3. 当原始值为 +1 或-1 且预嵌入秘密信息为 0 时,将这个位置归 0 并视为无效,在下一个 DCT 系数上重新嵌入
|
||||||
|
|
||||||
|
* 编写代码实现嵌入,并观察 DCT 系数变化
|
||||||
|
[代码实现](F3.py)
|
||||||
|
|
||||||
|
```
|
||||||
|
JPEG的DCT系数
|
||||||
|
{0: 32939, 1: 15730, 2: 13427, 3: 11523, 4: 9540, 5: 7957, 6: 6607, 7: 5697, 8: 4834, -1: 15294, -2: 13637, -3: 11479, -4: 9683, -5: 7979, -6: 6878, -7: 5631, -8: 4871}
|
||||||
|
Jsteg begin writing!
|
||||||
|
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
|
||||||
|
经过信息隐藏后JPEG的DCT系数变化
|
||||||
|
{0: 32939, 1: 15730, 2: 12552, 3: 12398, 4: 8739, 5: 8758, 6: 6165, 7: 6139, 8: 4487, -1: 15294, -2: 12721, -3: 12395, -4: 8891, -5: 8771, -6: 6319, -7: 6190, -8: 4463}
|
||||||
|
F3steg begin writing!
|
||||||
|
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
|
||||||
|
经过信息隐藏后JPEG的DCT系数变化
|
||||||
|
{0: 47068, 1: 13416, 2: 13519, 3: 10075, 4: 9545, 5: 7077, 6: 6650, 7: 5016, 8: 4754, -1: 13308, -2: 13668, -3: 10124, -4: 9571, -5: 7249, -6: 6591, -7: 5098, -8: 4733}
|
||||||
|
F4steg begin writing!
|
||||||
|
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
|
||||||
|
经过信息隐藏后JPEG的DCT系数变化
|
||||||
|
{0: 59320, 1: 13618, 2: 11987, 3: 9875, 4: 8328, 5: 6860, 6: 5883, 7: 4910, 8: 4239, -1: 13692, -2: 11976, -3: 9976, -4: 8428, -5: 7007, -6: 5834, -7: 4964, -8: 4190}
|
||||||
|
```
|
||||||
|
|
||||||
|
* 条形图绘制
|
||||||
|
|
||||||
|

|
||||||
|
|
||||||
|
* 未经过信息隐藏的 DCT 系数,系数近似符合拉普拉斯分布,具有几个典型特点
|
||||||
|
* 对称性 以 0 为中心达到最大值,两侧分布近似对称
|
||||||
|
* 单侧单调性 以 0 值为中心达到最大值,两侧单调下降
|
||||||
|
* 梯度下降性 小值样点较多,大值样点较少,分布曲线在两侧下降梯度逐渐减小
|
||||||
|

|
||||||
|
* JSteg 隐写的 DCT 系数
|
||||||
|
* JSteg 隐写可嵌入信息的 DCT 系数较少,隐写量较小,且相邻数值样点的个数接近,如 2 和 3,-2 和-3 形成了值对,卡方特征变化明显,因而提出了 F3 隐写
|
||||||
|

|
||||||
|
* F3 隐写的 DCT 系数
|
||||||
|
* F3 的设计虽然防止了相邻值出现数量接近的现象,也维持了分布函数的对称性,但使得偶数的分布增加,没有满足单调性
|
||||||
|
* 这是因为载体绝对值为 1 的数值较多,当其被修改为 0 时,嵌入算法继续嵌入直到找到一个偶数值,或者将一个奇数值改为偶数值,这样绝对值为 1 的系数可以支持嵌入 1,但是不支持嵌入 0,需要使用或制造一个偶数
|
||||||
|
* 另外,0 系数的数量有相应的增加,产生分布曲线向 0 收缩的现象
|
||||||
|
|
||||||
|
### F4 隐写
|
||||||
|
|
||||||
|
* 为了克服 F3 的缺陷,F4 对不同正负号的奇偶系数采用了不同的嵌入与消息表示方法
|
||||||
|
* **F4 用负偶数、正奇数代表嵌入了消息比特 1,用负奇数、正偶数代表嵌入了 0**
|
||||||
|
* 但仍然通过减小绝对值的方法进行修改,如果减小绝对值后系数为 0 则继续往下嵌入当前比特
|
||||||
|
* [代码实现](F4.py)
|
||||||
|

|
||||||
|
* F4 隐写的 DCT 系数
|
||||||
|
* F4 显然保持了载体分布函数的对称性,也保持了载体分布函数的单调性与梯度下降性
|
||||||
|
* 但 F4 依然存在使含密载体分布函数形状向 0 收缩的现象
|
||||||
|
|
||||||
|
### F5 隐写
|
||||||
|
|
||||||
|
F5 隐写实现了基于汉明码的矩阵编码隐写,在一个分组上最多修改 R=1 次以嵌入 $2^r-1-r$ 比特,采用的基本嵌入方法是基于 F4 隐写的
|
||||||
|
|
||||||
|
F5 的嵌入步骤
|
||||||
|
|
||||||
|
- 获得嵌入域。若输入的是位图,则进行 JPEG 编码得到 JPEG 系数;若输入的是 JPEG 图像,则进行熵编码的解码得到 JPEG 系数
|
||||||
|
- 位置置乱。根据口令生成的密钥位一个伪随机数发生器,基于伪随机数发生器置乱 JPEG 系数的位置
|
||||||
|
- 编码参数确定。为了提高嵌入效率,一般希望 n 尽可能大,因此,根据载体中可用系数的数量与消息的长度确定参数 r,并计算$n=2^r-1$
|
||||||
|
- 基于($n=2^r-1,r$)的汉明分组码得到编码校验矩阵,开始嵌入消息:
|
||||||
|
- ① 按置乱后的顺序取下面 n 个非零系数,在其中的 LSB 序列中按照以上编码嵌入 n-r 比特的消息;
|
||||||
|
- ② 如果未发生修改,并且还有需要嵌入的消息,则返回 ① 继续嵌入下一分组;
|
||||||
|
- ③ 如果进行了修改,则判断是不是有系数值收缩到 0,如果没有,并且还有需要嵌入的消息则返回 ① 继续嵌入下一分组,如果有,取出一个新的非零系数组成新的一组 n 个非零系数,在其中的 LSB 序列中按照以上编码重新嵌入以上 n-r 比特的消息,直到没有修改或收缩,最后,如果还有需要嵌入的消息,则返回 ① 继续嵌入下一分组
|
||||||
|
- 位置逆置乱。恢复 DCT 系数原来的位置顺序
|
||||||
|
- 熵编码。按照 JPEG 标准无损压缩 DCT 量化系数,得到 JPEG 文件
|
||||||
|
|
||||||
|
# 参考资料
|
||||||
|
|
||||||
|
* 隐写学原理与技术 By 赵险峰
|
||||||
|
* 数字媒体中的隐写术 By J.Fridrich
|
||||||
|
|
|
||||||
|
|
@ -0,0 +1,118 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Wed Sep 18 19:59:14 2019
|
||||||
|
|
||||||
|
@author: Lee
|
||||||
|
"""
|
||||||
|
|
||||||
|
import os
|
||||||
|
import sys
|
||||||
|
os.environ['CUDA_DEVICE_ORDER'] = 'PCI_BUS_ID'
|
||||||
|
os.environ['CUDA_VISIBLE_DEVICES'] = '1' # set a GPU (with GPU Number)
|
||||||
|
home = os.path.expanduser("~")
|
||||||
|
sys.path.append(home + '/tflib/') # path for 'tflib' folder
|
||||||
|
import matplotlib.pyplot as plt
|
||||||
|
from scipy.io import loadmat
|
||||||
|
from SCA_SRNet_Spatial import * # use 'SCA_SRNet_JPEG' for JPEG domain
|
||||||
|
|
||||||
|
|
||||||
|
def trnGen(cover_path, stego_path, cover_beta_path, stego_beta_path, thread_idx=0, n_threads=1):
|
||||||
|
IL=os.listdir(cover_path)
|
||||||
|
img_shape = plt.imread(cover_path +IL[0]).shape
|
||||||
|
batch = np.empty((2, img_shape[0], img_shape[1], 2), dtype='float32')
|
||||||
|
while True:
|
||||||
|
indx = np.random.permutation(len(IL))
|
||||||
|
for i in indx:
|
||||||
|
batch[0,:,:,0] = plt.imread(cover_path + IL[i]) # use loadmat for loading JPEG decompressed images
|
||||||
|
batch[0,:,:,1] = loadmat(cover_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images
|
||||||
|
batch[1,:,:,0] = plt.imread(stego_path + IL[i]) # use loadmat for loading JPEG decompressed images
|
||||||
|
batch[1,:,:,1] = loadmat(stego_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images
|
||||||
|
rot = random.randint(0,3)
|
||||||
|
if rand() < 0.5:
|
||||||
|
yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]
|
||||||
|
else:
|
||||||
|
yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')]
|
||||||
|
|
||||||
|
|
||||||
|
def valGen(cover_path, stego_path, cover_beta_path, stego_beta_path, thread_idx=0, n_threads=1):
|
||||||
|
IL=os.listdir(cover_path)
|
||||||
|
img_shape = plt.imread(cover_path +IL[0]).shape
|
||||||
|
batch = np.empty((2, img_shape[0], img_shape[1], 2), dtype='float32')
|
||||||
|
while True:
|
||||||
|
for i in range(len(IL)):
|
||||||
|
batch[0,:,:,0] = plt.imread(cover_path + IL[i]) # use loadmat for loading JPEG decompressed images
|
||||||
|
batch[0,:,:,1] = loadmat(cover_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images
|
||||||
|
batch[1,:,:,0] = plt.imread(stego_path + IL[i]) # use loadmat for loading JPEG decompressed images
|
||||||
|
batch[1,:,:,1] = loadmat(stego_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images
|
||||||
|
yield [batch, np.array([0,1], dtype='uint8') ]
|
||||||
|
|
||||||
|
|
||||||
|
train_batch_size = 32
|
||||||
|
valid_batch_size = 40
|
||||||
|
max_iter = 500000
|
||||||
|
train_interval=100
|
||||||
|
valid_interval=5000
|
||||||
|
save_interval=5000
|
||||||
|
num_runner_threads=10
|
||||||
|
|
||||||
|
# save Betas as '.mat' files with variable name "Beta" and put them in thier corresponding directoroies. Make sure
|
||||||
|
# all mat files in the directories can be loaded in Python without any errors.
|
||||||
|
|
||||||
|
TRAIN_COVER_DIR = '/media/Cover_TRN/'
|
||||||
|
TRAIN_STEGO_DIR = '/media/Stego_WOW_0.5_TRN/'
|
||||||
|
TRAIN_COVER_BETA_DIR = '/media/Beta_Cover_WOW_0.5_TRN/'
|
||||||
|
TRAIN_STEGO_BETA_DIR = '/media/Beta_Stego_WOW_0.5_TRN/'
|
||||||
|
|
||||||
|
VALID_COVER_DIR = '/media/Cover_VAL/'
|
||||||
|
VALID_STEGO_DIR = '/media/Stego_WOW_0.5_VAL/'
|
||||||
|
VALID_COVER_BETA_DIR = '/media/Beta_Cover_WOW_0.5_VAL/'
|
||||||
|
VALID_STEGO_BETA_DIR = '/media/Beta_Stego_WOW_0.5_VAL/'
|
||||||
|
|
||||||
|
train_gen = partial(trnGen, \
|
||||||
|
TRAIN_COVER_DIR, TRAIN_STEGO_DIR, TRAIN_COVER_BETA_DIR, TRAIN_STEGO_BETA_DIR)
|
||||||
|
valid_gen = partial(valGen, \
|
||||||
|
VALID_COVER_DIR, VALID_STEGO_DIR, VALID_COVER_BETA_DIR, VALID_STEGO_BETA_DIR)
|
||||||
|
|
||||||
|
LOG_DIR= '/media/LogFiles/SCA_WOW_0.5' # path for a log direcotry
|
||||||
|
# load_path= LOG_DIR + 'Model_460000.ckpt' # continue training from a specific checkpoint
|
||||||
|
load_path=None # training from scratch
|
||||||
|
|
||||||
|
if not os.path.exists(LOG_DIR):
|
||||||
|
os.makedirs(LOG_DIR)
|
||||||
|
|
||||||
|
train_ds_size = len(glob(TRAIN_COVER_DIR + '/*')) * 2
|
||||||
|
valid_ds_size = len(glob(VALID_COVER_DIR +'/*')) * 2
|
||||||
|
print 'train_ds_size: %i'%train_ds_size
|
||||||
|
print 'valid_ds_size: %i'%valid_ds_size
|
||||||
|
|
||||||
|
if valid_ds_size % valid_batch_size != 0:
|
||||||
|
raise ValueError("change batch size for validation")
|
||||||
|
|
||||||
|
optimizer = AdamaxOptimizer
|
||||||
|
boundaries = [400000] # learning rate adjustment at iteration 400K
|
||||||
|
values = [0.001, 0.0001] # learning rates
|
||||||
|
train(SCA_SRNet, train_gen, valid_gen , train_batch_size, valid_batch_size, valid_ds_size, \
|
||||||
|
optimizer, boundaries, values, train_interval, valid_interval, max_iter,\
|
||||||
|
save_interval, LOG_DIR,num_runner_threads, load_path)
|
||||||
|
|
||||||
|
|
||||||
|
# Testing
|
||||||
|
TEST_COVER_DIR = '/media/Cover_TST/'
|
||||||
|
TEST_STEGO_DIR = '/media/Stego_WOW_0.5_TST/'
|
||||||
|
TEST_COVER_BETA_DIR = '/media/Beta_Cover_WOW_0.5_TST/'
|
||||||
|
TEST_STEGO_BETA_DIR = '/media/Beta_Stego_WOW_0.5_TST/'
|
||||||
|
|
||||||
|
test_batch_size=40
|
||||||
|
LOG_DIR = '/media/LogFiles/SCA_WOW_0.5/'
|
||||||
|
LOAD_DIR = LOG_DIR + 'Model_435000.ckpt' # loading from a specific checkpoint
|
||||||
|
|
||||||
|
test_gen = partial(gen_valid, \
|
||||||
|
TEST_COVER_DIR, TEST_STEGO_DIR)
|
||||||
|
|
||||||
|
test_ds_size = len(glob(TEST_COVER_DIR + '/*')) * 2
|
||||||
|
print 'test_ds_size: %i'%test_ds_size
|
||||||
|
|
||||||
|
if test_ds_size % test_batch_size != 0:
|
||||||
|
raise ValueError("change batch size for testing!")
|
||||||
|
|
||||||
|
test_dataset(SCA_SRNet, test_gen, test_batch_size, test_ds_size, LOAD_DIR)
|
||||||
|
|
@ -0,0 +1,178 @@
|
||||||
|
{
|
||||||
|
"cells": [
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"import os\n",
|
||||||
|
"import sys\n",
|
||||||
|
"os.environ['CUDA_DEVICE_ORDER'] = 'PCI_BUS_ID'\n",
|
||||||
|
"os.environ['CUDA_VISIBLE_DEVICES'] = '1' # set a GPU (with GPU Number)\n",
|
||||||
|
"home = os.path.expanduser(\"~\")\n",
|
||||||
|
"sys.path.append(home + '/tflib/') # path for 'tflib' folder\n",
|
||||||
|
"import matplotlib.pyplot as plt\n",
|
||||||
|
"from scipy.io import loadmat\n",
|
||||||
|
"from SCA_SRNet_Spatial import * # use 'SCA_SRNet_JPEG' for JPEG domain"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"def trnGen(cover_path, stego_path, cover_beta_path, stego_beta_path, thread_idx=0, n_threads=1):\n",
|
||||||
|
" IL=os.listdir(cover_path)\n",
|
||||||
|
" img_shape = plt.imread(cover_path +IL[0]).shape\n",
|
||||||
|
" batch = np.empty((2, img_shape[0], img_shape[1], 2), dtype='float32')\n",
|
||||||
|
" while True:\n",
|
||||||
|
" indx = np.random.permutation(len(IL))\n",
|
||||||
|
" for i in indx:\n",
|
||||||
|
" batch[0,:,:,0] = plt.imread(cover_path + IL[i]) # use loadmat for loading JPEG decompressed images \n",
|
||||||
|
" batch[0,:,:,1] = loadmat(cover_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images\n",
|
||||||
|
" batch[1,:,:,0] = plt.imread(stego_path + IL[i]) # use loadmat for loading JPEG decompressed images \n",
|
||||||
|
" batch[1,:,:,1] = loadmat(stego_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images\n",
|
||||||
|
" rot = random.randint(0,3)\n",
|
||||||
|
" if rand() < 0.5:\n",
|
||||||
|
" yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]\n",
|
||||||
|
" else:\n",
|
||||||
|
" yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')] "
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"def valGen(cover_path, stego_path, cover_beta_path, stego_beta_path, thread_idx=0, n_threads=1):\n",
|
||||||
|
" IL=os.listdir(cover_path)\n",
|
||||||
|
" img_shape = plt.imread(cover_path +IL[0]).shape\n",
|
||||||
|
" batch = np.empty((2, img_shape[0], img_shape[1], 2), dtype='float32')\n",
|
||||||
|
" while True:\n",
|
||||||
|
" for i in range(len(IL)):\n",
|
||||||
|
" batch[0,:,:,0] = plt.imread(cover_path + IL[i]) # use loadmat for loading JPEG decompressed images \n",
|
||||||
|
" batch[0,:,:,1] = loadmat(cover_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images\n",
|
||||||
|
" batch[1,:,:,0] = plt.imread(stego_path + IL[i]) # use loadmat for loading JPEG decompressed images \n",
|
||||||
|
" batch[1,:,:,1] = loadmat(stego_beta_path + IL[i].replace('pgm','mat'))['Beta'] # adjust for JPEG images\n",
|
||||||
|
" yield [batch, np.array([0,1], dtype='uint8') ]"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"train_batch_size = 32\n",
|
||||||
|
"valid_batch_size = 40\n",
|
||||||
|
"max_iter = 500000\n",
|
||||||
|
"train_interval=100\n",
|
||||||
|
"valid_interval=5000\n",
|
||||||
|
"save_interval=5000\n",
|
||||||
|
"num_runner_threads=10\n",
|
||||||
|
"\n",
|
||||||
|
"# save Betas as '.mat' files with variable name \"Beta\" and put them in thier corresponding directoroies. Make sure \n",
|
||||||
|
"# all mat files in the directories can be loaded in Python without any errors.\n",
|
||||||
|
"\n",
|
||||||
|
"TRAIN_COVER_DIR = '/media/Cover_TRN/'\n",
|
||||||
|
"TRAIN_STEGO_DIR = '/media/Stego_WOW_0.5_TRN/'\n",
|
||||||
|
"TRAIN_COVER_BETA_DIR = '/media/Beta_Cover_WOW_0.5_TRN/'\n",
|
||||||
|
"TRAIN_STEGO_BETA_DIR = '/media/Beta_Stego_WOW_0.5_TRN/'\n",
|
||||||
|
"\n",
|
||||||
|
"VALID_COVER_DIR = '/media/Cover_VAL/'\n",
|
||||||
|
"VALID_STEGO_DIR = '/media/Stego_WOW_0.5_VAL/'\n",
|
||||||
|
"VALID_COVER_BETA_DIR = '/media/Beta_Cover_WOW_0.5_VAL/'\n",
|
||||||
|
"VALID_STEGO_BETA_DIR = '/media/Beta_Stego_WOW_0.5_VAL/'\n",
|
||||||
|
"\n",
|
||||||
|
"train_gen = partial(trnGen, \\\n",
|
||||||
|
" TRAIN_COVER_DIR, TRAIN_STEGO_DIR, TRAIN_COVER_BETA_DIR, TRAIN_STEGO_BETA_DIR) \n",
|
||||||
|
"valid_gen = partial(valGen, \\\n",
|
||||||
|
" VALID_COVER_DIR, VALID_STEGO_DIR, VALID_COVER_BETA_DIR, VALID_STEGO_BETA_DIR)\n",
|
||||||
|
"\n",
|
||||||
|
"LOG_DIR= '/media/LogFiles/SCA_WOW_0.5' # path for a log direcotry\n",
|
||||||
|
"# load_path= LOG_DIR + 'Model_460000.ckpt' # continue training from a specific checkpoint\n",
|
||||||
|
"load_path=None # training from scratch\n",
|
||||||
|
"\n",
|
||||||
|
"if not os.path.exists(LOG_DIR):\n",
|
||||||
|
" os.makedirs(LOG_DIR)\n",
|
||||||
|
" \n",
|
||||||
|
"train_ds_size = len(glob(TRAIN_COVER_DIR + '/*')) * 2\n",
|
||||||
|
"valid_ds_size = len(glob(VALID_COVER_DIR +'/*')) * 2\n",
|
||||||
|
"print 'train_ds_size: %i'%train_ds_size\n",
|
||||||
|
"print 'valid_ds_size: %i'%valid_ds_size\n",
|
||||||
|
"\n",
|
||||||
|
"if valid_ds_size % valid_batch_size != 0:\n",
|
||||||
|
" raise ValueError(\"change batch size for validation\")\n",
|
||||||
|
"\n",
|
||||||
|
"optimizer = AdamaxOptimizer\n",
|
||||||
|
"boundaries = [400000] # learning rate adjustment at iteration 400K\n",
|
||||||
|
"values = [0.001, 0.0001] # learning rates\n",
|
||||||
|
"train(SCA_SRNet, train_gen, valid_gen , train_batch_size, valid_batch_size, valid_ds_size, \\\n",
|
||||||
|
" optimizer, boundaries, values, train_interval, valid_interval, max_iter,\\\n",
|
||||||
|
" save_interval, LOG_DIR,num_runner_threads, load_path)"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"# Testing \n",
|
||||||
|
"TEST_COVER_DIR = '/media/Cover_TST/'\n",
|
||||||
|
"TEST_STEGO_DIR = '/media/Stego_WOW_0.5_TST/'\n",
|
||||||
|
"TEST_COVER_BETA_DIR = '/media/Beta_Cover_WOW_0.5_TST/'\n",
|
||||||
|
"TEST_STEGO_BETA_DIR = '/media/Beta_Stego_WOW_0.5_TST/'\n",
|
||||||
|
"\n",
|
||||||
|
"test_batch_size=40\n",
|
||||||
|
"LOG_DIR = '/media/LogFiles/SCA_WOW_0.5/' \n",
|
||||||
|
"LOAD_DIR = LOG_DIR + 'Model_435000.ckpt' # loading from a specific checkpoint\n",
|
||||||
|
"\n",
|
||||||
|
"test_gen = partial(gen_valid, \\\n",
|
||||||
|
" TEST_COVER_DIR, TEST_STEGO_DIR)\n",
|
||||||
|
"\n",
|
||||||
|
"test_ds_size = len(glob(TEST_COVER_DIR + '/*')) * 2\n",
|
||||||
|
"print 'test_ds_size: %i'%test_ds_size\n",
|
||||||
|
"\n",
|
||||||
|
"if test_ds_size % test_batch_size != 0:\n",
|
||||||
|
" raise ValueError(\"change batch size for testing!\")\n",
|
||||||
|
"\n",
|
||||||
|
"test_dataset(SCA_SRNet, test_gen, test_batch_size, test_ds_size, LOAD_DIR)"
|
||||||
|
]
|
||||||
|
}
|
||||||
|
],
|
||||||
|
"metadata": {
|
||||||
|
"kernelspec": {
|
||||||
|
"display_name": "Python 2",
|
||||||
|
"language": "python",
|
||||||
|
"name": "python2"
|
||||||
|
},
|
||||||
|
"language_info": {
|
||||||
|
"codemirror_mode": {
|
||||||
|
"name": "ipython",
|
||||||
|
"version": 2
|
||||||
|
},
|
||||||
|
"file_extension": ".py",
|
||||||
|
"mimetype": "text/x-python",
|
||||||
|
"name": "python",
|
||||||
|
"nbconvert_exporter": "python",
|
||||||
|
"pygments_lexer": "ipython2",
|
||||||
|
"version": "2.7.15rc1"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"nbformat": 4,
|
||||||
|
"nbformat_minor": 2
|
||||||
|
}
|
||||||
|
|
@ -0,0 +1,127 @@
|
||||||
|
{
|
||||||
|
"cells": [
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"import os\n",
|
||||||
|
"import sys\n",
|
||||||
|
"os.environ['CUDA_DEVICE_ORDER'] = 'PCI_BUS_ID'\n",
|
||||||
|
"os.environ['CUDA_VISIBLE_DEVICES'] = '1' # set a GPU (with GPU Number)\n",
|
||||||
|
"home = os.path.expanduser(\"~\")\n",
|
||||||
|
"sys.path.append(home + '/tflib/') # path for 'tflib' folder\n",
|
||||||
|
"from SRNet import *"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"train_batch_size = 32\n",
|
||||||
|
"valid_batch_size = 40\n",
|
||||||
|
"max_iter = 500000\n",
|
||||||
|
"train_interval=100\n",
|
||||||
|
"valid_interval=5000\n",
|
||||||
|
"save_interval=5000\n",
|
||||||
|
"num_runner_threads=10\n",
|
||||||
|
"\n",
|
||||||
|
"# Cover and Stego directories for training and validation. For the spatial domain put cover and stego images in their \n",
|
||||||
|
"# corresponding direcotries. For the JPEG domain, decompress images to the spatial domain without rounding to integers and \n",
|
||||||
|
"# save them as '.mat' files with variable name \"im\". Put the '.mat' files in thier corresponding directoroies. Make sure \n",
|
||||||
|
"# all mat files in the directories can be loaded in Python without any errors.\n",
|
||||||
|
"\n",
|
||||||
|
"TRAIN_COVER_DIR = '/media/TRN/Cover/'\n",
|
||||||
|
"TRAIN_STEGO_DIR = '/media/TRN/JUNI_75_04/'\n",
|
||||||
|
"\n",
|
||||||
|
"VALID_COVER_DIR = '/media/VAL/Cover/'\n",
|
||||||
|
"VALID_STEGO_DIR = '/media/VAL/JUNI_75_04/'\n",
|
||||||
|
" \n",
|
||||||
|
"train_gen = partial(gen_flip_and_rot, \\\n",
|
||||||
|
" TRAIN_COVER_DIR, TRAIN_STEGO_DIR ) \n",
|
||||||
|
"valid_gen = partial(gen_valid, \\\n",
|
||||||
|
" VALID_COVER_DIR, VALID_STEGO_DIR)\n",
|
||||||
|
"\n",
|
||||||
|
"LOG_DIR = '/media/LogFiles/JUNI_75_04' # path for a log direcotry \n",
|
||||||
|
"# load_path = LOG_DIR + 'Model_460000.ckpt' # continue training from a specific checkpoint\n",
|
||||||
|
"load_path=None # training from scratch\n",
|
||||||
|
"\n",
|
||||||
|
"if not os.path.exists(LOG_DIR):\n",
|
||||||
|
" os.makedirs(LOG_DIR)\n",
|
||||||
|
"\n",
|
||||||
|
"train_ds_size = len(glob(TRAIN_COVER_DIR + '/*')) * 2\n",
|
||||||
|
"valid_ds_size = len(glob(VALID_COVER_DIR +'/*')) * 2\n",
|
||||||
|
"print 'train_ds_size: %i'%train_ds_size\n",
|
||||||
|
"print 'valid_ds_size: %i'%valid_ds_size\n",
|
||||||
|
"\n",
|
||||||
|
"if valid_ds_size % valid_batch_size != 0:\n",
|
||||||
|
" raise ValueError(\"change batch size for validation\")\n",
|
||||||
|
" \n",
|
||||||
|
"optimizer = AdamaxOptimizer\n",
|
||||||
|
"boundaries = [400000] # learning rate adjustment at iteration 400K\n",
|
||||||
|
"values = [0.001, 0.0001] # learning rates\n",
|
||||||
|
"\n",
|
||||||
|
"train(SRNet, train_gen, valid_gen , train_batch_size, valid_batch_size, valid_ds_size, \\\n",
|
||||||
|
" optimizer, boundaries, values, train_interval, valid_interval, max_iter,\\\n",
|
||||||
|
" save_interval, LOG_DIR,num_runner_threads, load_path)"
|
||||||
|
]
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"cell_type": "code",
|
||||||
|
"execution_count": null,
|
||||||
|
"metadata": {
|
||||||
|
"collapsed": true
|
||||||
|
},
|
||||||
|
"outputs": [],
|
||||||
|
"source": [
|
||||||
|
"# Testing \n",
|
||||||
|
"# Cover and Stego directories for testing\n",
|
||||||
|
"TEST_COVER_DIR = '/media/TST/Cover/'\n",
|
||||||
|
"TEST_STEGO_DIR = '/media/TST/JUNI_75_04/'\n",
|
||||||
|
"\n",
|
||||||
|
"test_batch_size=40\n",
|
||||||
|
"LOG_DIR = '/media/LogFiles/JUNI_75_04/' \n",
|
||||||
|
"LOAD_CKPT = LOG_DIR + 'Model_435000.ckpt' # loading from a specific checkpoint\n",
|
||||||
|
"\n",
|
||||||
|
"test_gen = partial(gen_valid, \\\n",
|
||||||
|
" TEST_COVER_DIR, TEST_STEGO_DIR)\n",
|
||||||
|
"\n",
|
||||||
|
"test_ds_size = len(glob(TEST_COVER_DIR + '/*')) * 2\n",
|
||||||
|
"print 'test_ds_size: %i'%test_ds_size\n",
|
||||||
|
"\n",
|
||||||
|
"if test_ds_size % test_batch_size != 0:\n",
|
||||||
|
" raise ValueError(\"change batch size for testing!\")\n",
|
||||||
|
"\n",
|
||||||
|
"test_dataset(SRNet, test_gen, test_batch_size, test_ds_size, LOAD_CKPT)"
|
||||||
|
]
|
||||||
|
}
|
||||||
|
],
|
||||||
|
"metadata": {
|
||||||
|
"kernelspec": {
|
||||||
|
"display_name": "Python 2",
|
||||||
|
"language": "python",
|
||||||
|
"name": "python2"
|
||||||
|
},
|
||||||
|
"language_info": {
|
||||||
|
"codemirror_mode": {
|
||||||
|
"name": "ipython",
|
||||||
|
"version": 2
|
||||||
|
},
|
||||||
|
"file_extension": ".py",
|
||||||
|
"mimetype": "text/x-python",
|
||||||
|
"name": "python",
|
||||||
|
"nbconvert_exporter": "python",
|
||||||
|
"pygments_lexer": "ipython2",
|
||||||
|
"version": "2.7.15rc1"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"nbformat": 4,
|
||||||
|
"nbformat_minor": 2
|
||||||
|
}
|
||||||
|
|
@ -0,0 +1,133 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
from functools import partial
|
||||||
|
from tensorflow.contrib import layers
|
||||||
|
from tensorflow.contrib.framework import arg_scope
|
||||||
|
import functools
|
||||||
|
from queues import *
|
||||||
|
from generator import *
|
||||||
|
from utils_multistep_lr import *
|
||||||
|
|
||||||
|
class SCA_SRNet(Model):
|
||||||
|
def _build_model(self, input_batch):
|
||||||
|
inputs_image, inputs_Beta = tf.split(input_batch, num_or_size_splits=2, axis=3)
|
||||||
|
if self.data_format == 'NCHW':
|
||||||
|
reduction_axis = [2,3]
|
||||||
|
_inputs_image = tf.cast(tf.transpose(inputs_image, [0, 3, 1, 2]), tf.float32)
|
||||||
|
_inputs_Beta = tf.cast(tf.transpose(inputs_Beta, [0, 3, 1, 2]), tf.float32)
|
||||||
|
else:
|
||||||
|
reduction_axis = [1,2]
|
||||||
|
_inputs_image = tf.cast(inputs_image, tf.float32)
|
||||||
|
_inputs_Beta = tf.cast(inputs_Beta, tf.float32)
|
||||||
|
with arg_scope([layers.conv2d], num_outputs=16,
|
||||||
|
kernel_size=3, stride=1, padding='SAME',
|
||||||
|
data_format=self.data_format,
|
||||||
|
activation_fn=None,
|
||||||
|
weights_initializer=layers.variance_scaling_initializer(),
|
||||||
|
weights_regularizer=layers.l2_regularizer(2e-4),
|
||||||
|
biases_initializer=tf.constant_initializer(0.2),
|
||||||
|
biases_regularizer=None),\
|
||||||
|
arg_scope([layers.batch_norm],
|
||||||
|
decay=0.9, center=True, scale=True,
|
||||||
|
updates_collections=None, is_training=self.is_training,
|
||||||
|
fused=True, data_format=self.data_format),\
|
||||||
|
arg_scope([layers.avg_pool2d],
|
||||||
|
kernel_size=[3,3], stride=[2,2], padding='SAME',
|
||||||
|
data_format=self.data_format):
|
||||||
|
with tf.variable_scope('Layer1'): # 256*256
|
||||||
|
W = tf.get_variable('W', shape=[3,3,1,64],\
|
||||||
|
initializer=layers.variance_scaling_initializer(), \
|
||||||
|
dtype=tf.float32, \
|
||||||
|
regularizer=layers.l2_regularizer(5e-4))
|
||||||
|
b = tf.get_variable('b', shape=[64], dtype=tf.float32, \
|
||||||
|
initializer=tf.constant_initializer(0.2))
|
||||||
|
conv = tf.nn.bias_add( \
|
||||||
|
tf.nn.conv2d(tf.cast(_inputs_image, tf.float32), \
|
||||||
|
W, [1,1,1,1], 'SAME', \
|
||||||
|
data_format=self.data_format), b, \
|
||||||
|
data_format=self.data_format, name='Layer1')
|
||||||
|
actv=tf.nn.relu(conv)
|
||||||
|
prob_map = tf.sqrt(tf.nn.conv2d(tf.cast(_inputs_Beta, tf.float32), \
|
||||||
|
tf.abs(W), [1,1,1,1], 'SAME', \
|
||||||
|
data_format=self.data_format))
|
||||||
|
out_L1=tf.add_n([actv,prob_map])
|
||||||
|
with tf.variable_scope('Layer2'): # 256*256
|
||||||
|
conv=layers.conv2d(out_L1)
|
||||||
|
actv=tf.nn.relu(layers.batch_norm(conv))
|
||||||
|
with tf.variable_scope('Layer3'): # 256*256
|
||||||
|
conv1=layers.conv2d(actv)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn2=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(actv, bn2)
|
||||||
|
with tf.variable_scope('Layer4'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn2=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn2)
|
||||||
|
with tf.variable_scope('Layer5'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer6'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer7'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer8'): # 256*256
|
||||||
|
convs = layers.conv2d(res, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer9'): # 128*128
|
||||||
|
convs = layers.conv2d(res, num_outputs=64, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=64)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=64)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer10'): # 64*64
|
||||||
|
convs = layers.conv2d(res, num_outputs=128, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=128)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=128)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer11'): # 32*32
|
||||||
|
convs = layers.conv2d(res, num_outputs=256, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=256)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=256)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer12'): # 16*16
|
||||||
|
conv1=layers.conv2d(res, num_outputs=512)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=512)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
avgp = tf.reduce_mean(bn, reduction_axis, keep_dims=True )
|
||||||
|
ip=layers.fully_connected(layers.flatten(avgp), num_outputs=2,
|
||||||
|
activation_fn=None, normalizer_fn=None,
|
||||||
|
weights_initializer=tf.random_normal_initializer(mean=0., stddev=0.01),
|
||||||
|
biases_initializer=tf.constant_initializer(0.), scope='ip')
|
||||||
|
self.outputs = ip
|
||||||
|
return self.outputs
|
||||||
|
|
@ -0,0 +1,133 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
from functools import partial
|
||||||
|
from tensorflow.contrib import layers
|
||||||
|
from tensorflow.contrib.framework import arg_scope
|
||||||
|
import functools
|
||||||
|
from tflib.queues import *
|
||||||
|
from tflib.generator import *
|
||||||
|
from tflib.utils_multistep_lr import *
|
||||||
|
|
||||||
|
class SCA_SRNet(Model):
|
||||||
|
def _build_model(self, input_batch):
|
||||||
|
inputs_image, inputs_Beta = tf.split(input_batch, num_or_size_splits=2, axis=3)
|
||||||
|
if self.data_format == 'NCHW':
|
||||||
|
reduction_axis = [2,3]
|
||||||
|
_inputs_image = tf.cast(tf.transpose(inputs_image, [0, 3, 1, 2]), tf.float32)
|
||||||
|
_inputs_Beta = tf.cast(tf.transpose(inputs_Beta, [0, 3, 1, 2]), tf.float32)
|
||||||
|
else:
|
||||||
|
reduction_axis = [1,2]
|
||||||
|
_inputs_image = tf.cast(inputs_image, tf.float32)
|
||||||
|
_inputs_Beta = tf.cast(inputs_Beta, tf.float32)
|
||||||
|
with arg_scope([layers.conv2d], num_outputs=16,
|
||||||
|
kernel_size=3, stride=1, padding='SAME',
|
||||||
|
data_format=self.data_format,
|
||||||
|
activation_fn=None,
|
||||||
|
weights_initializer=layers.variance_scaling_initializer(),
|
||||||
|
weights_regularizer=layers.l2_regularizer(2e-4),
|
||||||
|
biases_initializer=tf.constant_initializer(0.2),
|
||||||
|
biases_regularizer=None),\
|
||||||
|
arg_scope([layers.batch_norm],
|
||||||
|
decay=0.9, center=True, scale=True,
|
||||||
|
updates_collections=None, is_training=self.is_training,
|
||||||
|
fused=True, data_format=self.data_format),\
|
||||||
|
arg_scope([layers.avg_pool2d],
|
||||||
|
kernel_size=[3,3], stride=[2,2], padding='SAME',
|
||||||
|
data_format=self.data_format):
|
||||||
|
with tf.variable_scope('Layer1'): # 256*256
|
||||||
|
W = tf.get_variable('W', shape=[3,3,1,64],\
|
||||||
|
initializer=layers.variance_scaling_initializer(), \
|
||||||
|
dtype=tf.float32, \
|
||||||
|
regularizer=layers.l2_regularizer(5e-4))
|
||||||
|
b = tf.get_variable('b', shape=[64], dtype=tf.float32, \
|
||||||
|
initializer=tf.constant_initializer(0.2))
|
||||||
|
conv = tf.nn.bias_add( \
|
||||||
|
tf.nn.conv2d(tf.cast(_inputs_image, tf.float32), \
|
||||||
|
W, [1,1,1,1], 'SAME', \
|
||||||
|
data_format=self.data_format), b, \
|
||||||
|
data_format=self.data_format, name='Layer1')
|
||||||
|
actv=tf.nn.relu(conv)
|
||||||
|
prob_map = tf.nn.conv2d(tf.cast(_inputs_Beta, tf.float32), \
|
||||||
|
tf.abs(W), [1,1,1,1], 'SAME', \
|
||||||
|
data_format=self.data_format)
|
||||||
|
out_L1=tf.add_n([actv,prob_map])
|
||||||
|
with tf.variable_scope('Layer2'): # 256*256
|
||||||
|
conv=layers.conv2d(out_L1)
|
||||||
|
actv=tf.nn.relu(layers.batch_norm(conv))
|
||||||
|
with tf.variable_scope('Layer3'): # 256*256
|
||||||
|
conv1=layers.conv2d(actv)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn2=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(actv, bn2)
|
||||||
|
with tf.variable_scope('Layer4'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn2=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn2)
|
||||||
|
with tf.variable_scope('Layer5'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer6'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer7'): # 256*256
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer8'): # 256*256
|
||||||
|
convs = layers.conv2d(res, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer9'): # 128*128
|
||||||
|
convs = layers.conv2d(res, num_outputs=64, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=64)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=64)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer10'): # 64*64
|
||||||
|
convs = layers.conv2d(res, num_outputs=128, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=128)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=128)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer11'): # 32*32
|
||||||
|
convs = layers.conv2d(res, num_outputs=256, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=256)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=256)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer12'): # 16*16
|
||||||
|
conv1=layers.conv2d(res, num_outputs=512)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=512)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
avgp = tf.reduce_mean(bn, reduction_axis, keep_dims=True )
|
||||||
|
ip=layers.fully_connected(layers.flatten(avgp), num_outputs=2,
|
||||||
|
activation_fn=None, normalizer_fn=None,
|
||||||
|
weights_initializer=tf.random_normal_initializer(mean=0., stddev=0.01),
|
||||||
|
biases_initializer=tf.constant_initializer(0.), scope='ip')
|
||||||
|
self.outputs = ip
|
||||||
|
return self.outputs
|
||||||
|
|
@ -0,0 +1,117 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
from functools import partial
|
||||||
|
from tensorflow.contrib import layers
|
||||||
|
from tensorflow.contrib.framework import arg_scope
|
||||||
|
import functools
|
||||||
|
from tflib.queues import *
|
||||||
|
from tflib.generator import *
|
||||||
|
from tflib.utils_multistep_lr import *
|
||||||
|
|
||||||
|
class SRNet(Model):
|
||||||
|
def _build_model(self, inputs):
|
||||||
|
self.inputs = inputs
|
||||||
|
if self.data_format == 'NCHW':
|
||||||
|
reduction_axis = [2,3]
|
||||||
|
_inputs = tf.cast(tf.transpose(inputs, [0, 3, 1, 2]), tf.float32)
|
||||||
|
else:
|
||||||
|
reduction_axis = [1,2]
|
||||||
|
_inputs = tf.cast(inputs, tf.float32)
|
||||||
|
with arg_scope([layers.conv2d], num_outputs=16,
|
||||||
|
kernel_size=3, stride=1, padding='SAME',
|
||||||
|
data_format=self.data_format,
|
||||||
|
activation_fn=None,
|
||||||
|
weights_initializer=layers.variance_scaling_initializer(),
|
||||||
|
weights_regularizer=layers.l2_regularizer(2e-4),
|
||||||
|
biases_initializer=tf.constant_initializer(0.2),
|
||||||
|
biases_regularizer=None),\
|
||||||
|
arg_scope([layers.batch_norm],
|
||||||
|
decay=0.9, center=True, scale=True,
|
||||||
|
updates_collections=None, is_training=self.is_training,
|
||||||
|
fused=True, data_format=self.data_format),\
|
||||||
|
arg_scope([layers.avg_pool2d],
|
||||||
|
kernel_size=[3,3], stride=[2,2], padding='SAME',
|
||||||
|
data_format=self.data_format):
|
||||||
|
with tf.variable_scope('Layer1'):
|
||||||
|
conv=layers.conv2d(_inputs, num_outputs=64, kernel_size=3)
|
||||||
|
actv=tf.nn.relu(layers.batch_norm(conv))
|
||||||
|
with tf.variable_scope('Layer2'):
|
||||||
|
conv=layers.conv2d(actv)
|
||||||
|
actv=tf.nn.relu(layers.batch_norm(conv))
|
||||||
|
with tf.variable_scope('Layer3'):
|
||||||
|
conv1=layers.conv2d(actv)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn2=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(actv, bn2)
|
||||||
|
with tf.variable_scope('Layer4'):
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn2=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn2)
|
||||||
|
with tf.variable_scope('Layer5'):
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer6'):
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer7'):
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
res= tf.add(res, bn)
|
||||||
|
with tf.variable_scope('Layer8'):
|
||||||
|
convs = layers.conv2d(res, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer9'):
|
||||||
|
convs = layers.conv2d(res, num_outputs=64, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=64)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=64)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer10'):
|
||||||
|
convs = layers.conv2d(res, num_outputs=128, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=128)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=128)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer11'):
|
||||||
|
convs = layers.conv2d(res, num_outputs=256, kernel_size=1, stride=2)
|
||||||
|
convs = layers.batch_norm(convs)
|
||||||
|
conv1=layers.conv2d(res, num_outputs=256)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=256)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
pool = layers.avg_pool2d(bn)
|
||||||
|
res= tf.add(convs, pool)
|
||||||
|
with tf.variable_scope('Layer12'):
|
||||||
|
conv1=layers.conv2d(res, num_outputs=512)
|
||||||
|
actv1=tf.nn.relu(layers.batch_norm(conv1))
|
||||||
|
conv2=layers.conv2d(actv1, num_outputs=512)
|
||||||
|
bn=layers.batch_norm(conv2)
|
||||||
|
avgp = tf.reduce_mean(bn, reduction_axis, keep_dims=True )
|
||||||
|
ip=layers.fully_connected(layers.flatten(avgp), num_outputs=2,
|
||||||
|
activation_fn=None, normalizer_fn=None,
|
||||||
|
weights_initializer=tf.random_normal_initializer(mean=0., stddev=0.01),
|
||||||
|
biases_initializer=tf.constant_initializer(0.), scope='ip')
|
||||||
|
self.outputs = ip
|
||||||
|
return self.outputs
|
||||||
|
|
@ -0,0 +1,81 @@
|
||||||
|
import numpy as np
|
||||||
|
from scipy import misc, io
|
||||||
|
from glob import glob
|
||||||
|
import random
|
||||||
|
# from itertools import izip
|
||||||
|
from random import random as rand
|
||||||
|
from random import shuffle
|
||||||
|
|
||||||
|
def gen_flip_and_rot(cover_dir, stego_dir, thread_idx, n_threads):
|
||||||
|
cover_list = sorted(glob(cover_dir + '/*'))
|
||||||
|
# print(cover_list)
|
||||||
|
stego_list = sorted(glob(stego_dir + '/*'))
|
||||||
|
nb_data = len(cover_list)
|
||||||
|
assert len(stego_list) != 0, "the stego directory '%s' is empty" % stego_dir
|
||||||
|
assert nb_data != 0, "the cover directory '%s' is empty" % cover_dir
|
||||||
|
assert len(stego_list) == nb_data, "the cover directory and " + \
|
||||||
|
"the stego directory don't " + \
|
||||||
|
"have the same number of files " + \
|
||||||
|
"respectively %d and %d" % (nb_data, + \
|
||||||
|
len(stego_list))
|
||||||
|
load_mat=cover_list[0].endswith('.mat')
|
||||||
|
if load_mat:
|
||||||
|
img = io.loadmat(cover_list[0])['im']
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2,img_shape[0],img_shape[1],1), dtype='float32')
|
||||||
|
else:
|
||||||
|
img = misc.imread(cover_list[0])
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2,img_shape[0],img_shape[1],1), dtype='uint8')
|
||||||
|
|
||||||
|
iterable = list(zip(cover_list, stego_list))
|
||||||
|
while True:
|
||||||
|
shuffle(iterable)
|
||||||
|
print("shuffling……")
|
||||||
|
for cover_path, stego_path in iterable:
|
||||||
|
if load_mat:
|
||||||
|
batch[0,:,:,0] = io.loadmat(cover_path)['im']
|
||||||
|
batch[1,:,:,0] = io.loadmat(stego_path)['im']
|
||||||
|
else:
|
||||||
|
batch[0,:,:,0] = misc.imread(cover_path)
|
||||||
|
batch[1,:,:,0] = misc.imread(stego_path)
|
||||||
|
rot = random.randint(0,3)
|
||||||
|
if rand() < 0.5:
|
||||||
|
yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]
|
||||||
|
else:
|
||||||
|
yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')]
|
||||||
|
|
||||||
|
|
||||||
|
def gen_valid(cover_dir, stego_dir, thread_idx, n_threads):
|
||||||
|
cover_list = sorted(glob(cover_dir + '/*'))
|
||||||
|
stego_list = sorted(glob(stego_dir + '/*'))
|
||||||
|
nb_data = len(cover_list)
|
||||||
|
assert len(stego_list) != 0, "the stego directory '%s' is empty" % stego_dir
|
||||||
|
assert nb_data != 0, "the cover directory '%s' is empty" % cover_dir
|
||||||
|
assert len(stego_list) == nb_data, "the cover directory and " + \
|
||||||
|
"the stego directory don't " + \
|
||||||
|
"have the same number of files " + \
|
||||||
|
"respectively %d and %d" % (nb_data, \
|
||||||
|
len(stego_list))
|
||||||
|
load_mat=cover_list[0].endswith('.mat')
|
||||||
|
if load_mat:
|
||||||
|
img = io.loadmat(cover_list[0])['im']
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2,img_shape[0],img_shape[1],1), dtype='float32')
|
||||||
|
else:
|
||||||
|
img = misc.imread(cover_list[0])
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2,img_shape[0],img_shape[1],1), dtype='uint8')
|
||||||
|
img_shape = img.shape
|
||||||
|
|
||||||
|
labels = np.array([0, 1], dtype='uint8')
|
||||||
|
while True:
|
||||||
|
for cover_path, stego_path in zip(cover_list, stego_list):
|
||||||
|
if load_mat:
|
||||||
|
batch[0,:,:,0] = io.loadmat(cover_path)['im']
|
||||||
|
batch[1,:,:,0] = io.loadmat(stego_path)['im']
|
||||||
|
else:
|
||||||
|
batch[0,:,:,0] = misc.imread(cover_path)
|
||||||
|
batch[1,:,:,0] = misc.imread(stego_path)
|
||||||
|
# print([batch,labels])
|
||||||
|
yield [batch, labels]
|
||||||
|
|
@ -0,0 +1,68 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
import threading
|
||||||
|
|
||||||
|
class GeneratorRunner():
|
||||||
|
"""
|
||||||
|
This class manage a multithreaded queue filled with a generator
|
||||||
|
"""
|
||||||
|
def __init__(self, generator, capacity):
|
||||||
|
"""
|
||||||
|
inputs: generator feeding the data, must have thread_idx
|
||||||
|
as parameter (but the parameter may be not used)
|
||||||
|
"""
|
||||||
|
self.generator = generator
|
||||||
|
_input = generator(0,1).__next__()
|
||||||
|
if type(_input) is not list:
|
||||||
|
raise ValueError("generator doesn't return" \
|
||||||
|
"a list: %r" % type(_input))
|
||||||
|
input_batch_size = _input[0].shape[0]
|
||||||
|
if not all(_input[i].shape[0] == input_batch_size for i in range(len(_input))):
|
||||||
|
raise ValueError("all the inputs doesn't have the same batch size,"\
|
||||||
|
"the batch sizes are: %s" % [_input[i].shape[0] for i in range(len(_input))])
|
||||||
|
self.data = []
|
||||||
|
self.dtypes = []
|
||||||
|
self.shapes = []
|
||||||
|
for i in range(len(_input)):
|
||||||
|
self.shapes.append(_input[i].shape[1:])
|
||||||
|
self.dtypes.append(_input[i].dtype)
|
||||||
|
self.data.append(tf.placeholder(dtype=self.dtypes[i], \
|
||||||
|
shape=(input_batch_size,) + self.shapes[i]))
|
||||||
|
self.queue = tf.FIFOQueue(capacity, shapes=self.shapes, \
|
||||||
|
dtypes=self.dtypes)
|
||||||
|
self.enqueue_op = self.queue.enqueue_many(self.data)
|
||||||
|
self.close_queue_op = self.queue.close(cancel_pending_enqueues=True)
|
||||||
|
|
||||||
|
def get_batched_inputs(self, batch_size):
|
||||||
|
"""
|
||||||
|
Return tensors containing a batch of generated data
|
||||||
|
"""
|
||||||
|
batch = self.queue.dequeue_many(batch_size)
|
||||||
|
return batch
|
||||||
|
|
||||||
|
def thread_main(self, sess, thread_idx=0, n_threads=1):
|
||||||
|
try:
|
||||||
|
for data in self.generator(thread_idx, n_threads):
|
||||||
|
sess.run(self.enqueue_op, feed_dict={i: d \
|
||||||
|
for i, d in zip(self.data, data)})
|
||||||
|
if self.stop_threads:
|
||||||
|
return
|
||||||
|
except RuntimeError:
|
||||||
|
pass
|
||||||
|
|
||||||
|
def start_threads(self, sess, n_threads=1):
|
||||||
|
self.stop_threads = False
|
||||||
|
self.threads = []
|
||||||
|
for n in range(n_threads):
|
||||||
|
t = threading.Thread(target=self.thread_main, args=(sess, n, n_threads))
|
||||||
|
t.daemon = True
|
||||||
|
t.start()
|
||||||
|
self.threads.append(t)
|
||||||
|
return self.threads
|
||||||
|
|
||||||
|
def stop_runner(self, sess):
|
||||||
|
self.stop_threads = True
|
||||||
|
sess.run(self.close_queue_op)
|
||||||
|
|
||||||
|
def queueSelection(runners, sel, batch_size):
|
||||||
|
selection_queue = tf.FIFOQueue.from_list(sel, [r.queue for r in runners])
|
||||||
|
return selection_queue.dequeue_many(batch_size)
|
||||||
|
|
@ -0,0 +1,231 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
import time
|
||||||
|
from tflib.queues import *
|
||||||
|
from tflib.generator import *
|
||||||
|
|
||||||
|
class average_summary:
|
||||||
|
def __init__(self, variable, name, num_iterations):
|
||||||
|
self.sum_variable = tf.get_variable(name, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0), \
|
||||||
|
dtype=variable.dtype.base_dtype, \
|
||||||
|
trainable=False, \
|
||||||
|
collections=[tf.GraphKeys.LOCAL_VARIABLES])
|
||||||
|
with tf.control_dependencies([variable]):
|
||||||
|
self.increment_op = tf.assign_add(self.sum_variable, variable)
|
||||||
|
self.mean_variable = self.sum_variable / float(num_iterations)
|
||||||
|
self.summary = tf.summary.scalar(name, self.mean_variable)
|
||||||
|
with tf.control_dependencies([self.summary]):
|
||||||
|
self.reset_variable_op = tf.assign(self.sum_variable, 0)
|
||||||
|
|
||||||
|
def add_summary(self, sess, writer, step):
|
||||||
|
s, _ = sess.run([self.summary, self.reset_variable_op])
|
||||||
|
writer.add_summary(s, step)
|
||||||
|
|
||||||
|
class Model:
|
||||||
|
def __init__(self, is_training=None, data_format='NCHW'):
|
||||||
|
self.data_format = data_format
|
||||||
|
if is_training is None:
|
||||||
|
self.is_training = tf.get_variable('is_training', dtype=tf.bool, \
|
||||||
|
initializer=tf.constant_initializer(True), \
|
||||||
|
trainable=False)
|
||||||
|
else:
|
||||||
|
self.is_training = is_training
|
||||||
|
|
||||||
|
def _build_model(self, inputs):
|
||||||
|
raise NotImplementedError('Here is your model definition')
|
||||||
|
|
||||||
|
def _build_losses(self, labels):
|
||||||
|
self.labels = tf.cast(labels, tf.int64)
|
||||||
|
with tf.variable_scope('loss'):
|
||||||
|
oh = tf.one_hot(self.labels, 2)
|
||||||
|
xen_loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits( \
|
||||||
|
labels=oh,logits=self.outputs))
|
||||||
|
reg_losses = tf.get_collection(tf.GraphKeys.REGULARIZATION_LOSSES)
|
||||||
|
self.loss = tf.add_n([xen_loss] + reg_losses)
|
||||||
|
with tf.variable_scope('accuracy'):
|
||||||
|
am = tf.argmax(self.outputs, 1)
|
||||||
|
equal = tf.equal(am, self.labels)
|
||||||
|
self.accuracy = tf.reduce_mean(tf.cast(equal, tf.float32))
|
||||||
|
return self.loss, self.accuracy
|
||||||
|
|
||||||
|
def train(model_class, train_gen, valid_gen, train_batch_size, \
|
||||||
|
valid_batch_size, valid_ds_size, optimizer, boundaries, values, \
|
||||||
|
train_interval, valid_interval, max_iter, \
|
||||||
|
save_interval, log_path, num_runner_threads=1, \
|
||||||
|
load_path=None):
|
||||||
|
tf.reset_default_graph()
|
||||||
|
train_runner = GeneratorRunner(train_gen, train_batch_size * 10)
|
||||||
|
valid_runner = GeneratorRunner(valid_gen, valid_batch_size * 10)
|
||||||
|
is_training = tf.get_variable('is_training', dtype=tf.bool, \
|
||||||
|
initializer=True, trainable=False)
|
||||||
|
if train_batch_size == valid_batch_size:
|
||||||
|
batch_size = train_batch_size
|
||||||
|
disable_training_op = tf.assign(is_training, False)
|
||||||
|
enable_training_op = tf.assign(is_training, True)
|
||||||
|
else:
|
||||||
|
batch_size = tf.get_variable('batch_size', dtype=tf.int32, \
|
||||||
|
initializer=train_batch_size, \
|
||||||
|
trainable=False, \
|
||||||
|
collections=[tf.GraphKeys.LOCAL_VARIABLES])
|
||||||
|
disable_training_op = tf.group(tf.assign(is_training, False), \
|
||||||
|
tf.assign(batch_size, valid_batch_size))
|
||||||
|
enable_training_op = tf.group(tf.assign(is_training, True), \
|
||||||
|
tf.assign(batch_size, train_batch_size))
|
||||||
|
img_batch, label_batch = queueSelection([valid_runner, train_runner], \
|
||||||
|
tf.cast(is_training, tf.int32), \
|
||||||
|
batch_size)
|
||||||
|
model = model_class(is_training, 'NCHW')
|
||||||
|
model._build_model(img_batch)
|
||||||
|
loss, accuracy = model._build_losses(label_batch)
|
||||||
|
train_loss_s = average_summary(loss, 'train_loss', train_interval)
|
||||||
|
train_accuracy_s = average_summary(accuracy, 'train_accuracy', \
|
||||||
|
train_interval)
|
||||||
|
valid_loss_s = average_summary(loss, 'valid_loss', \
|
||||||
|
float(valid_ds_size) / float(valid_batch_size))
|
||||||
|
valid_accuracy_s = average_summary(accuracy, 'valid_accuracy', \
|
||||||
|
float(valid_ds_size) / float(valid_batch_size))
|
||||||
|
global_step = tf.get_variable('global_step', dtype=tf.int32, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0), \
|
||||||
|
trainable=False)
|
||||||
|
|
||||||
|
learning_rate = tf.train.piecewise_constant(global_step, boundaries, values)
|
||||||
|
lr_summary = tf.summary.scalar('learning_rate', learning_rate)
|
||||||
|
optimizer = optimizer(learning_rate)
|
||||||
|
|
||||||
|
minimize_op = optimizer.minimize(loss, global_step)
|
||||||
|
train_op = tf.group(minimize_op, train_loss_s.increment_op, \
|
||||||
|
train_accuracy_s.increment_op)
|
||||||
|
increment_valid = tf.group(valid_loss_s.increment_op, \
|
||||||
|
valid_accuracy_s.increment_op)
|
||||||
|
init_op = tf.group(tf.global_variables_initializer(), \
|
||||||
|
tf.local_variables_initializer())
|
||||||
|
saver = tf.train.Saver(max_to_keep=10000)
|
||||||
|
with tf.Session() as sess:
|
||||||
|
sess.run(init_op)
|
||||||
|
if load_path is not None:
|
||||||
|
saver.restore(sess, load_path)
|
||||||
|
train_runner.start_threads(sess, num_runner_threads)
|
||||||
|
valid_runner.start_threads(sess, 1)
|
||||||
|
writer = tf.summary.FileWriter(log_path + '/LogFile/', \
|
||||||
|
sess.graph)
|
||||||
|
start = sess.run(global_step)
|
||||||
|
sess.run(disable_training_op)
|
||||||
|
sess.run([valid_loss_s.reset_variable_op, \
|
||||||
|
valid_accuracy_s.reset_variable_op, \
|
||||||
|
train_loss_s.reset_variable_op, \
|
||||||
|
train_accuracy_s.reset_variable_op])
|
||||||
|
_time = time.time()
|
||||||
|
for j in range(0, valid_ds_size, valid_batch_size):
|
||||||
|
sess.run([increment_valid])
|
||||||
|
_acc_val = sess.run(valid_accuracy_s.mean_variable)
|
||||||
|
print ("initial accuracy on validation set:", _acc_val)
|
||||||
|
print ("evaluation time on validation set:", time.time() - _time, "seconds")
|
||||||
|
valid_accuracy_s.add_summary(sess, writer, start)
|
||||||
|
valid_loss_s.add_summary(sess, writer, start)
|
||||||
|
sess.run(enable_training_op)
|
||||||
|
print ("network will be evaluatd every %i iterations on validation set" %valid_interval)
|
||||||
|
for i in range(start+1, max_iter+1):
|
||||||
|
sess.run(train_op)
|
||||||
|
if i % train_interval == 0:
|
||||||
|
train_loss_s.add_summary(sess, writer, i)
|
||||||
|
train_accuracy_s.add_summary(sess, writer, i)
|
||||||
|
s = sess.run(lr_summary)
|
||||||
|
writer.add_summary(s, i)
|
||||||
|
if i % valid_interval == 0:
|
||||||
|
sess.run(disable_training_op)
|
||||||
|
for j in range(0, valid_ds_size, valid_batch_size):
|
||||||
|
sess.run([increment_valid])
|
||||||
|
valid_loss_s.add_summary(sess, writer, i)
|
||||||
|
valid_accuracy_s.add_summary(sess, writer, i)
|
||||||
|
sess.run(enable_training_op)
|
||||||
|
if i % save_interval == 0:
|
||||||
|
saver.save(sess, log_path + '/Model_' + str(i) + '.ckpt')
|
||||||
|
|
||||||
|
def test_dataset(model_class, gen, batch_size, ds_size, load_path):
|
||||||
|
tf.reset_default_graph()
|
||||||
|
runner = GeneratorRunner(gen, batch_size * 10)
|
||||||
|
img_batch, label_batch = runner.get_batched_inputs(batch_size)
|
||||||
|
model = model_class(False, 'NCHW')
|
||||||
|
model._build_model(img_batch)
|
||||||
|
loss, accuracy = model._build_losses(label_batch)
|
||||||
|
loss_summary = average_summary(loss, 'loss', \
|
||||||
|
float(ds_size) / float(batch_size))
|
||||||
|
accuracy_summary = average_summary(accuracy, 'accuracy', \
|
||||||
|
float(ds_size) / float(batch_size))
|
||||||
|
increment_op = tf.group(loss_summary.increment_op, \
|
||||||
|
accuracy_summary.increment_op)
|
||||||
|
global_step = tf.get_variable('global_step', dtype=tf.int32, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0), \
|
||||||
|
trainable=False)
|
||||||
|
init_op = tf.group(tf.global_variables_initializer(), \
|
||||||
|
tf.local_variables_initializer())
|
||||||
|
saver = tf.train.Saver(max_to_keep=10000)
|
||||||
|
with tf.Session() as sess:
|
||||||
|
sess.run(init_op)
|
||||||
|
saver.restore(sess, load_path)
|
||||||
|
runner.start_threads(sess, 1)
|
||||||
|
for j in range(0, ds_size, batch_size):
|
||||||
|
sess.run(increment_op)
|
||||||
|
mean_loss, mean_accuracy = sess.run([loss_summary.mean_variable ,\
|
||||||
|
accuracy_summary.mean_variable])
|
||||||
|
print ("Accuracy:", mean_accuracy, " | Loss:", mean_loss)
|
||||||
|
|
||||||
|
### Implementation of Adamax optimizer, taken from : https://github.com/openai/iaf/blob/master/tf_utils/adamax.py
|
||||||
|
from tensorflow.python.ops import control_flow_ops
|
||||||
|
from tensorflow.python.ops import math_ops
|
||||||
|
from tensorflow.python.ops import state_ops
|
||||||
|
from tensorflow.python.framework import ops
|
||||||
|
from tensorflow.python.training import optimizer
|
||||||
|
import tensorflow as tf
|
||||||
|
|
||||||
|
class AdamaxOptimizer(optimizer.Optimizer):
|
||||||
|
"""
|
||||||
|
Optimizer that implements the Adamax algorithm.
|
||||||
|
See [Kingma et. al., 2014](http://arxiv.org/abs/1412.6980)
|
||||||
|
([pdf](http://arxiv.org/pdf/1412.6980.pdf)).
|
||||||
|
@@__init__
|
||||||
|
"""
|
||||||
|
|
||||||
|
def __init__(self, learning_rate=0.001, beta1=0.9, beta2=0.999, use_locking=False, name="Adamax"):
|
||||||
|
super(AdamaxOptimizer, self).__init__(use_locking, name)
|
||||||
|
self._lr = learning_rate
|
||||||
|
self._beta1 = beta1
|
||||||
|
self._beta2 = beta2
|
||||||
|
|
||||||
|
# Tensor versions of the constructor arguments, created in _prepare().
|
||||||
|
self._lr_t = None
|
||||||
|
self._beta1_t = None
|
||||||
|
self._beta2_t = None
|
||||||
|
|
||||||
|
def _prepare(self):
|
||||||
|
self._lr_t = ops.convert_to_tensor(self._lr, name="learning_rate")
|
||||||
|
self._beta1_t = ops.convert_to_tensor(self._beta1, name="beta1")
|
||||||
|
self._beta2_t = ops.convert_to_tensor(self._beta2, name="beta2")
|
||||||
|
|
||||||
|
def _create_slots(self, var_list):
|
||||||
|
# Create slots for the first and second moments.
|
||||||
|
for v in var_list:
|
||||||
|
self._zeros_slot(v, "m", self._name)
|
||||||
|
self._zeros_slot(v, "v", self._name)
|
||||||
|
|
||||||
|
def _apply_dense(self, grad, var):
|
||||||
|
lr_t = math_ops.cast(self._lr_t, var.dtype.base_dtype)
|
||||||
|
beta1_t = math_ops.cast(self._beta1_t, var.dtype.base_dtype)
|
||||||
|
beta2_t = math_ops.cast(self._beta2_t, var.dtype.base_dtype)
|
||||||
|
if var.dtype.base_dtype == tf.float16:
|
||||||
|
eps = 1e-7 # Can't use 1e-8 due to underflow -- not sure if it makes a big difference.
|
||||||
|
else:
|
||||||
|
eps = 1e-8
|
||||||
|
|
||||||
|
v = self.get_slot(var, "v")
|
||||||
|
v_t = v.assign(beta1_t * v + (1. - beta1_t) * grad)
|
||||||
|
m = self.get_slot(var, "m")
|
||||||
|
m_t = m.assign(tf.maximum(beta2_t * m + eps, tf.abs(grad)))
|
||||||
|
g_t = v_t / m_t
|
||||||
|
|
||||||
|
var_update = state_ops.assign_sub(var, lr_t * g_t)
|
||||||
|
return control_flow_ops.group(*[var_update, m_t, v_t])
|
||||||
|
|
||||||
|
def _apply_sparse(self, grad, var):
|
||||||
|
raise NotImplementedError("Sparse gradient updates are not supported.")
|
||||||
|
|
||||||
|
|
@ -0,0 +1,8 @@
|
||||||
|
# YeNet-Tensorflow
|
||||||
|
Tensorflow implementation of "Deep Learning Hierarchical Representation for Image Steganalysis" by Jian Ye, Jiangqun Ni and Yang Yi
|
||||||
|
|
||||||
|
## Dataset
|
||||||
|
Training and validation contains there own cover and stego / beta maps images directory, stego images must have the same name than there corresponding cover
|
||||||
|
|
||||||
|
## Publication
|
||||||
|
[The publication can be found here](http://ieeexplore.ieee.org/document/7937836/)
|
||||||
Binary file not shown.
|
|
@ -0,0 +1,119 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
from tensorflow.contrib import layers
|
||||||
|
from tensorflow.contrib.framework import add_arg_scope, arg_scope, arg_scoped_arguments
|
||||||
|
import layers as my_layers
|
||||||
|
from utils import *
|
||||||
|
|
||||||
|
SRM_Kernels = np.load('SRM_Kernels.npy')
|
||||||
|
|
||||||
|
class YeNet(Model):
|
||||||
|
def __init__(self, is_training=None, data_format='NCHW', \
|
||||||
|
with_bn=False, tlu_threshold=3):
|
||||||
|
super(YeNet, self).__init__(is_training=is_training, \
|
||||||
|
data_format=data_format)
|
||||||
|
self.with_bn = with_bn
|
||||||
|
self.tlu_threshold = tlu_threshold
|
||||||
|
|
||||||
|
def _build_model(self, inputs):
|
||||||
|
self.inputs = inputs
|
||||||
|
if self.data_format == 'NCHW':
|
||||||
|
channel_axis = 1
|
||||||
|
_inputs = tf.cast(tf.transpose(inputs, [0, 3, 1, 2]), tf.float32)
|
||||||
|
else:
|
||||||
|
channel_axis = 3
|
||||||
|
_inputs = tf.cast(inputs, tf.float32)
|
||||||
|
self.L = []
|
||||||
|
with arg_scope([layers.avg_pool2d], \
|
||||||
|
padding='VALID', data_format=self.data_format):
|
||||||
|
with tf.variable_scope('SRM_preprocess'):
|
||||||
|
W_SRM = tf.get_variable('W', initializer=SRM_Kernels, \
|
||||||
|
dtype=tf.float32, \
|
||||||
|
regularizer=None)
|
||||||
|
b = tf.get_variable('b', shape=[30], dtype=tf.float32, \
|
||||||
|
initializer=tf.constant_initializer(0.))
|
||||||
|
self.L.append(tf.nn.bias_add( \
|
||||||
|
tf.nn.conv2d(_inputs, \
|
||||||
|
W_SRM, [1,1,1,1], 'VALID', \
|
||||||
|
data_format=self.data_format), b, \
|
||||||
|
data_format=self.data_format, name='Layer1'))
|
||||||
|
self.L.append(tf.clip_by_value(self.L[-1], \
|
||||||
|
-self.tlu_threshold, self.tlu_threshold, \
|
||||||
|
name='TLU'))
|
||||||
|
with tf.variable_scope('ConvNetwork'):
|
||||||
|
with arg_scope([my_layers.conv2d], num_outputs=30, \
|
||||||
|
kernel_size=3, stride=1, padding='VALID', \
|
||||||
|
data_format=self.data_format, \
|
||||||
|
activation_fn=tf.nn.relu, \
|
||||||
|
weights_initializer=layers.xavier_initializer_conv2d(), \
|
||||||
|
weights_regularizer=layers.l2_regularizer(5e-4), \
|
||||||
|
biases_initializer=tf.constant_initializer(0.2), \
|
||||||
|
biases_regularizer=None), arg_scope([layers.batch_norm], \
|
||||||
|
decay=0.9, center=True, scale=True, \
|
||||||
|
updates_collections=None, is_training=self.is_training, \
|
||||||
|
fused=True, data_format=self.data_format):
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm1'))
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
scope='Layer2'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm2'))
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
scope='Layer3'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm3'))
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
scope='Layer4'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm4'))
|
||||||
|
self.L.append(layers.avg_pool2d(self.L[-1], \
|
||||||
|
kernel_size=[2,2], scope='Stride1'))
|
||||||
|
with arg_scope([my_layers.conv2d], kernel_size=5, \
|
||||||
|
num_outputs=32):
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
scope='Layer5'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm5'))
|
||||||
|
self.L.append(layers.avg_pool2d(self.L[-1], \
|
||||||
|
kernel_size=[3,3], \
|
||||||
|
scope='Stride2'))
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
scope='Layer6'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm6'))
|
||||||
|
self.L.append(layers.avg_pool2d(self.L[-1], \
|
||||||
|
kernel_size=[3,3], \
|
||||||
|
scope='Stride3'))
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
scope='Layer7'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm7'))
|
||||||
|
self.L.append(layers.avg_pool2d(self.L[-1], \
|
||||||
|
kernel_size=[3,3], \
|
||||||
|
scope='Stride4'))
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
num_outputs=16, \
|
||||||
|
scope='Layer8'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm8'))
|
||||||
|
self.L.append(my_layers.conv2d(self.L[-1], \
|
||||||
|
num_outputs=16, stride=3, \
|
||||||
|
scope='Layer9'))
|
||||||
|
if self.with_bn:
|
||||||
|
self.L.append(layers.batch_norm(self.L[-1], \
|
||||||
|
scope='Norm9'))
|
||||||
|
self.L.append(layers.flatten(self.L[-1]))
|
||||||
|
self.L.append(layers.fully_connected(self.L[-1], num_outputs=2, \
|
||||||
|
activation_fn=None, normalizer_fn=None, \
|
||||||
|
weights_initializer=tf.random_normal_initializer(mean=0., stddev=0.01), \
|
||||||
|
biases_initializer=tf.constant_initializer(0.), scope='ip'))
|
||||||
|
self.outputs = self.L[-1]
|
||||||
|
return self.outputs
|
||||||
|
|
||||||
Binary file not shown.
|
|
@ -0,0 +1,178 @@
|
||||||
|
import numpy as np
|
||||||
|
from scipy import misc, io
|
||||||
|
from glob import glob
|
||||||
|
import random
|
||||||
|
from itertools import izip
|
||||||
|
from random import random as rand
|
||||||
|
from random import shuffle
|
||||||
|
import h5py
|
||||||
|
|
||||||
|
def gen_embedding_otf(cover_dir, beta_dir, shuf_pair, \
|
||||||
|
thread_idx=0, n_threads=1):
|
||||||
|
cover_list = sorted(glob(cover_dir + '/*'))
|
||||||
|
beta_list = sorted(glob(beta_dir + '/*'))
|
||||||
|
nb_data = len(cover_list)
|
||||||
|
assert len(beta_list) != 0, "the beta directory '%s' is empty" % beta_dir
|
||||||
|
assert nb_data != 0, "the cover directory '%s' is empty" % cover_dir
|
||||||
|
assert len(beta_list) == nb_data, "the cover directory and " + \
|
||||||
|
"the beta directory don't " + \
|
||||||
|
"have the same number of files " + \
|
||||||
|
"respectively %d and %d" % (nb_data, \
|
||||||
|
len(beta_list))
|
||||||
|
img = misc.imread(cover_list[0])
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2, img_shape[0], img_shape[1], 1), dtype='uint8')
|
||||||
|
beta_map = np.empty(img_shape, dtype='<f8')
|
||||||
|
inf_map = np.empty(img_shape, dtype='bool')
|
||||||
|
rand_arr = np.empty(img_shape, dtype='float64')
|
||||||
|
shuf_cov = np.empty(img_shape, dtype='uint8')
|
||||||
|
while True:
|
||||||
|
if shuf_pair:
|
||||||
|
list_i = np.random.permutation(nb_data)
|
||||||
|
list_j = np.random.permutation(nb_data)
|
||||||
|
for i, j in izip(list_i, list_j):
|
||||||
|
batch[0,:,:,0] = misc.imread(cover_list[i])
|
||||||
|
beta_map[:,:] = io.loadmat(beta_list[j])['pChange']
|
||||||
|
shuf_cov[:,:] = misc.imread(cover_list[j])
|
||||||
|
rand_arr[:,:] = np.random.rand(img_shape[0], img_shape[1])
|
||||||
|
inf_map[:,:] = rand_arr < (beta_map / 2.)
|
||||||
|
batch[1,:,:,0] = np.copy(shuf_cov)
|
||||||
|
batch[1,np.logical_and(shuf_cov != 255, inf_map),0] += 1
|
||||||
|
batch[1,np.logical_and(shuf_cov != 0, \
|
||||||
|
np.logical_and(np.logical_not(inf_map), \
|
||||||
|
rand_arr < beta_map)), 0] -= 1
|
||||||
|
rot = random.randint(0,3)
|
||||||
|
if rand() < 0.5:
|
||||||
|
yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]
|
||||||
|
else:
|
||||||
|
yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')]
|
||||||
|
else:
|
||||||
|
list_i = np.random.permutation(nb_data)
|
||||||
|
for i in list_i:
|
||||||
|
batch[0,:,:,0] = misc.imread(cover_list[i])
|
||||||
|
beta_map[:,:] = io.loadmat(beta_list[i])['pChange']
|
||||||
|
rand_arr[:,:] = np.random.rand(img_shape[0], img_shape[1])
|
||||||
|
inf_map[:,:] = rand_arr < (beta_map / 2.)
|
||||||
|
batch[1,:,:,0] = np.copy(batch[0,:,:,0])
|
||||||
|
batch[1,np.logical_and(batch[0,:,:,0] != 255, inf_map),0] += 1
|
||||||
|
batch[1,np.logical_and(batch[0,:,:,0] != 0, \
|
||||||
|
np.logical_and(np.logical_not(inf_map), \
|
||||||
|
rand_arr < beta_map)), 0] -= 1
|
||||||
|
rot = random.randint(0,3)
|
||||||
|
if rand() < 0.5:
|
||||||
|
yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]
|
||||||
|
else:
|
||||||
|
yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')]
|
||||||
|
|
||||||
|
def gen_all_flip_and_rot(cover_dir, stego_dir, thread_idx, n_threads):
|
||||||
|
cover_list = sorted(glob(cover_dir + '/*'))
|
||||||
|
stego_list = sorted(glob(stego_dir + '/*'))
|
||||||
|
nb_data = len(cover_list)
|
||||||
|
assert len(stego_list) != 0, "the beta directory '%s' is empty" % stego_dir
|
||||||
|
assert nb_data != 0, "the cover directory '%s' is empty" % cover_dir
|
||||||
|
assert len(stego_list) == nb_data, "the cover directory and " + \
|
||||||
|
"the beta directory don't " + \
|
||||||
|
"have the same number of files " + \
|
||||||
|
"respectively %d and %d" % (nb_data, + \
|
||||||
|
len(stego_list))
|
||||||
|
img = misc.imread(cover_list[0])
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2,img_shape[0],img_shape[1],1), dtype='uint8')
|
||||||
|
iterable = zip(cover_list, stego_list)
|
||||||
|
for cover_path, stego_path in iterable:
|
||||||
|
batch[0,:,:,0] = misc.imread(cover_path)
|
||||||
|
batch[1,:,:,0] = misc.imread(stego_path)
|
||||||
|
for rot in range(4):
|
||||||
|
yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]
|
||||||
|
for rot in range(4):
|
||||||
|
yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')]
|
||||||
|
|
||||||
|
def gen_flip_and_rot(cover_dir, stego_dir, shuf_pair=False, thread_idx=0, n_threads=1):
|
||||||
|
cover_list = sorted(glob(cover_dir + '/*'))
|
||||||
|
stego_list = sorted(glob(stego_dir + '/*'))
|
||||||
|
nb_data = len(cover_list)
|
||||||
|
assert len(stego_list) != 0, "the beta directory '%s' is empty" % stego_dir
|
||||||
|
assert nb_data != 0, "the cover directory '%s' is empty" % cover_dir
|
||||||
|
assert len(stego_list) == nb_data, "the cover directory and " + \
|
||||||
|
"the beta directory don't " + \
|
||||||
|
"have the same number of files " + \
|
||||||
|
"respectively %d and %d" % (nb_data, + \
|
||||||
|
len(stego_list))
|
||||||
|
img = misc.imread(cover_list[0])
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2,img_shape[0],img_shape[1],1), dtype='uint8')
|
||||||
|
if not shuf_pair:
|
||||||
|
iterable = zip(cover_list, stego_list)
|
||||||
|
while True:
|
||||||
|
if shuf_pair:
|
||||||
|
shuffle(cover_list)
|
||||||
|
shuffle(stego_list)
|
||||||
|
iterable = izip(cover_list, stego_list)
|
||||||
|
else:
|
||||||
|
shuffle(iterable)
|
||||||
|
for cover_path, stego_path in iterable:
|
||||||
|
batch[0,:,:,0] = misc.imread(cover_path)
|
||||||
|
batch[1,:,:,0] = misc.imread(stego_path)
|
||||||
|
rot = random.randint(0,3)
|
||||||
|
if rand() < 0.5:
|
||||||
|
yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]
|
||||||
|
else:
|
||||||
|
yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')]
|
||||||
|
|
||||||
|
def gen_valid(cover_dir, stego_dir, thread_idx, n_threads):
|
||||||
|
cover_list = sorted(glob(cover_dir + '/*'))
|
||||||
|
stego_list = sorted(glob(stego_dir + '/*'))
|
||||||
|
nb_data = len(cover_list)
|
||||||
|
assert len(stego_list) != 0, "the beta directory '%s' is empty" % stego_dir
|
||||||
|
assert nb_data != 0, "the cover directory '%s' is empty" % cover_dir
|
||||||
|
assert len(stego_list) == nb_data, "the cover directory and " + \
|
||||||
|
"the beta directory don't " + \
|
||||||
|
"have the same number of files " + \
|
||||||
|
"respectively %d and %d" % (nb_data, \
|
||||||
|
len(stego_list))
|
||||||
|
img = misc.imread(cover_list[0])
|
||||||
|
img_shape = img.shape
|
||||||
|
batch = np.empty((2,img_shape[0],img_shape[1],1), dtype='uint8')
|
||||||
|
labels = np.array([0, 1], dtype='uint8')
|
||||||
|
while True:
|
||||||
|
for cover_path, stego_path in zip(cover_list, stego_list):
|
||||||
|
batch[0,:,:,0] = misc.imread(cover_path)
|
||||||
|
batch[1,:,:,0] = misc.imread(stego_path)
|
||||||
|
yield [batch, labels]
|
||||||
|
|
||||||
|
|
||||||
|
# def trainGen(thread_idx, n_threads):
|
||||||
|
# batch = np.empty((2,256,256,1), dtype='uint8')
|
||||||
|
# beta_map = np.empty((256, 256), dtype='<f8')
|
||||||
|
# inf_map = np.empty((256, 256), dtype='bool')
|
||||||
|
# rand_arr = np.empty((256, 256), dtype='float64')
|
||||||
|
|
||||||
|
# cover_ds_path = LOG_DIR + '/cover_train' + str(thread_idx) +'.txt'
|
||||||
|
# shuf_cover_ds_path = LOG_DIR + '/shuf_cover_train' + str(thread_idx) +'.txt'
|
||||||
|
# shuf_beta_ds_path = LOG_DIR + '/shuf_beta_train' + str(thread_idx) +'.txt'
|
||||||
|
# createSubFile(DS_PATH + 'train_cover.txt', \
|
||||||
|
# cover_ds_path, thread_idx, n_threads)
|
||||||
|
# createSubFile(DS_PATH + 'train_beta.txt', \
|
||||||
|
# shuf_beta_ds_path, thread_idx, n_threads)
|
||||||
|
# createSubFile(DS_PATH + 'train_cover.txt', \
|
||||||
|
# shuf_cover_ds_path, thread_idx, n_threads)
|
||||||
|
# while True:
|
||||||
|
# with open(cover_ds_path, 'r') as f_cover, \
|
||||||
|
# open(shuf_cover_ds_path, 'r') as f_shuf_cover, \
|
||||||
|
# open(shuf_beta_ds_path, 'r') as f_shuf_beta:
|
||||||
|
# for cov_path, shuf_cov_path, shuf_beta_path in \
|
||||||
|
# izip(f_cover, f_shuf_cover, f_shuf_beta):
|
||||||
|
# batch[0,:,:,0] = misc.imread(str.strip(cov_path))
|
||||||
|
# shuf_cov = misc.imread(str.strip(shuf_cov_path))
|
||||||
|
# beta_map[:,:] = io.loadmat(str.strip(shuf_beta_path))['pChange']
|
||||||
|
# rand_arr[:,:] = np.random.rand(256, 256)
|
||||||
|
# inf_map[:,:] = rand_arr < (beta_map / 2.)
|
||||||
|
# batch[1,:,:,0] = np.copy(shuf_cov)
|
||||||
|
# batch[1,np.logical_and(shuf_cov != 255, inf_map),0] += 1
|
||||||
|
# batch[1,np.logical_and(shuf_cov != 0, \
|
||||||
|
# np.logical_and(np.logical_not(inf_map), rand_arr < beta_map)), 0] -= 1
|
||||||
|
# rot = random.randint(0,3)
|
||||||
|
# if rand() < 0.5:
|
||||||
|
# yield [np.rot90(batch, rot, axes=[1,2]), np.array([0,1], dtype='uint8')]
|
||||||
|
# else:
|
||||||
|
# yield [np.flip(np.rot90(batch, rot, axes=[1,2]), axis=2), np.array([0,1], dtype='uint8')]
|
||||||
|
|
@ -0,0 +1,368 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
from tensorflow.contrib import layers
|
||||||
|
from tensorflow.contrib.framework import add_arg_scope
|
||||||
|
|
||||||
|
@add_arg_scope
|
||||||
|
def double_conv2d(ref_half, real_half,
|
||||||
|
num_outputs,
|
||||||
|
kernel_size,
|
||||||
|
stride=1,
|
||||||
|
padding='SAME',
|
||||||
|
data_format=None,
|
||||||
|
rate=1,
|
||||||
|
activation_fn=tf.nn.relu,
|
||||||
|
normalizer_fn=None,
|
||||||
|
normalize_after_activation=True,
|
||||||
|
normalizer_params=None,
|
||||||
|
weights_initializer=layers.xavier_initializer(),
|
||||||
|
weights_regularizer=None,
|
||||||
|
biases_initializer=tf.zeros_initializer(),
|
||||||
|
biases_regularizer=None,
|
||||||
|
reuse=None,
|
||||||
|
variables_collections=None,
|
||||||
|
outputs_collections=None,
|
||||||
|
trainable=True,
|
||||||
|
scope=None):
|
||||||
|
with tf.variable_scope(scope, 'Conv', reuse=reuse):
|
||||||
|
if data_format == 'NHWC':
|
||||||
|
num_inputs = real_half.get_shape().as_list()[3]
|
||||||
|
height = real_half.get_shape().as_list()[1]
|
||||||
|
width = real_half.get_shape().as_list()[2]
|
||||||
|
if isinstance(stride, int):
|
||||||
|
strides = [1, stride, stride, 1]
|
||||||
|
elif isinstance(stride, list) or isinstance(stride, tuple):
|
||||||
|
if len(stride) == 1:
|
||||||
|
strides = [1] + stride * 2 + [1]
|
||||||
|
else:
|
||||||
|
strides = [1, stride[0], stride[1], 1]
|
||||||
|
else:
|
||||||
|
raise TypeError('stride is not an int, list or' \
|
||||||
|
+ 'a tuple, is %s' % type(stride))
|
||||||
|
else:
|
||||||
|
num_inputs = real_half.get_shape().as_list()[1]
|
||||||
|
height = real_half.get_shape().as_list()[2]
|
||||||
|
width = real_half.get_shape().as_list()[3]
|
||||||
|
if isinstance(stride, int):
|
||||||
|
strides = [1, 1, stride, stride]
|
||||||
|
elif isinstance(stride, list) or isinstance(stride, tuple):
|
||||||
|
if len(stride) == 1:
|
||||||
|
strides = [1, 1] + stride * 2
|
||||||
|
else:
|
||||||
|
strides = [1, 1, stride[0], stride[1]]
|
||||||
|
else:
|
||||||
|
raise TypeError('stride is not an int, list or' \
|
||||||
|
+ 'a tuple, is %s' % type(stride))
|
||||||
|
if isinstance(kernel_size, int):
|
||||||
|
kernel_height = kernel_size
|
||||||
|
kernel_width = kernel_size
|
||||||
|
elif isinstance(kernel_size, list) \
|
||||||
|
or isinstance(kernel_size, tuple):
|
||||||
|
kernel_height = kernel_size[0]
|
||||||
|
kernel_width = kernel_size[1]
|
||||||
|
else:
|
||||||
|
raise ValueError('kernel_size is not an int, list or' \
|
||||||
|
+ 'a tuple, is %s' % type(kernel_size))
|
||||||
|
weights = tf.get_variable('weights', [kernel_height, \
|
||||||
|
kernel_width, num_inputs, num_outputs], \
|
||||||
|
'float32', weights_initializer, \
|
||||||
|
weights_regularizer, trainable, \
|
||||||
|
variables_collections)
|
||||||
|
ref_outputs = tf.nn.conv2d(ref_half, weights, strides, padding, \
|
||||||
|
data_format=data_format)
|
||||||
|
real_outputs = tf.nn.conv2d(real_half, weights, strides, padding, \
|
||||||
|
data_format=data_format)
|
||||||
|
if biases_initializer is not None:
|
||||||
|
biases = tf.get_variable('biases', [num_outputs], 'float32', \
|
||||||
|
biases_initializer, \
|
||||||
|
biases_regularizer, \
|
||||||
|
trainable, variables_collections)
|
||||||
|
ref_outputs = tf.nn.bias_add(ref_outputs, biases, data_format)
|
||||||
|
real_outputs = tf.nn.bias_add(real_outputs, biases, data_format)
|
||||||
|
if normalizer_fn is not None \
|
||||||
|
and not normalize_after_activation:
|
||||||
|
normalizer_params = normalizer_params or {}
|
||||||
|
ref_outputs, real_outputs = normalizer_fn(ref_outputs, \
|
||||||
|
real_outputs, \
|
||||||
|
**normalizer_params)
|
||||||
|
if activation_fn is not None:
|
||||||
|
ref_outputs = activation_fn(ref_outputs)
|
||||||
|
real_outputs = activation_fn(real_outputs)
|
||||||
|
if normalizer_fn is not None and normalize_after_activation:
|
||||||
|
normalizer_params = normalizer_params or {}
|
||||||
|
ref_outputs, real_outputs = normalizer_fn(ref_outputs, \
|
||||||
|
real_outputs,\
|
||||||
|
**normalizer_params)
|
||||||
|
return ref_outputs, real_outputs
|
||||||
|
|
||||||
|
@add_arg_scope
|
||||||
|
def conv2d(inputs,
|
||||||
|
num_outputs,
|
||||||
|
kernel_size,
|
||||||
|
stride=1,
|
||||||
|
padding='SAME',
|
||||||
|
data_format=None,
|
||||||
|
rate=1,
|
||||||
|
activation_fn=tf.nn.relu,
|
||||||
|
normalizer_fn=None,
|
||||||
|
normalize_after_activation=True,
|
||||||
|
normalizer_params=None,
|
||||||
|
weights_initializer=layers.xavier_initializer(),
|
||||||
|
weights_regularizer=None,
|
||||||
|
biases_initializer=tf.zeros_initializer(),
|
||||||
|
biases_regularizer=None,
|
||||||
|
reuse=None,
|
||||||
|
variables_collections=None,
|
||||||
|
outputs_collections=None,
|
||||||
|
trainable=True,
|
||||||
|
scope=None):
|
||||||
|
with tf.variable_scope(scope, 'Conv', reuse=reuse):
|
||||||
|
if data_format == 'NHWC':
|
||||||
|
num_inputs = inputs.get_shape().as_list()[3]
|
||||||
|
height = inputs.get_shape().as_list()[1]
|
||||||
|
width = inputs.get_shape().as_list()[2]
|
||||||
|
if isinstance(stride, int):
|
||||||
|
strides = [1, stride, stride, 1]
|
||||||
|
elif isinstance(stride, list) or isinstance(stride, tuple):
|
||||||
|
if len(stride) == 1:
|
||||||
|
strides = [1] + stride * 2 + [1]
|
||||||
|
else:
|
||||||
|
strides = [1, stride[0], stride[1], 1]
|
||||||
|
else:
|
||||||
|
raise TypeError('stride is not an int, list or' \
|
||||||
|
+ 'a tuple, is %s' % type(stride))
|
||||||
|
else:
|
||||||
|
num_inputs = inputs.get_shape().as_list()[1]
|
||||||
|
height = inputs.get_shape().as_list()[2]
|
||||||
|
width = inputs.get_shape().as_list()[3]
|
||||||
|
if isinstance(stride, int):
|
||||||
|
strides = [1, 1, stride, stride]
|
||||||
|
elif isinstance(stride, list) or isinstance(stride, tuple):
|
||||||
|
if len(stride) == 1:
|
||||||
|
strides = [1, 1] + stride * 2
|
||||||
|
else:
|
||||||
|
strides = [1, 1, stride[0], stride[1]]
|
||||||
|
else:
|
||||||
|
raise TypeError('stride is not an int, list or' \
|
||||||
|
+ 'a tuple, is %s' % type(stride))
|
||||||
|
if isinstance(kernel_size, int):
|
||||||
|
kernel_height = kernel_size
|
||||||
|
kernel_width = kernel_size
|
||||||
|
elif isinstance(kernel_size, list) \
|
||||||
|
or isinstance(kernel_size, tuple):
|
||||||
|
kernel_height = kernel_size[0]
|
||||||
|
kernel_width = kernel_size[1]
|
||||||
|
else:
|
||||||
|
raise ValueError('kernel_size is not an int, list or' \
|
||||||
|
+ 'a tuple, is %s' % type(kernel_size))
|
||||||
|
weights = tf.get_variable('weights', [kernel_height, \
|
||||||
|
kernel_width, num_inputs, num_outputs], \
|
||||||
|
'float32', weights_initializer, \
|
||||||
|
weights_regularizer, trainable, \
|
||||||
|
variables_collections)
|
||||||
|
outputs = tf.nn.conv2d(inputs, weights, strides, padding, \
|
||||||
|
data_format=data_format)
|
||||||
|
if biases_initializer is not None:
|
||||||
|
biases = tf.get_variable('biases', [num_outputs], 'float32', \
|
||||||
|
biases_initializer, \
|
||||||
|
biases_regularizer, \
|
||||||
|
trainable, variables_collections)
|
||||||
|
outputs = tf.nn.bias_add(outputs, biases, data_format)
|
||||||
|
if normalizer_fn is not None \
|
||||||
|
and not normalize_after_activation:
|
||||||
|
normalizer_params = normalizer_params or {}
|
||||||
|
outputs = normalizer_fn(outputs, **normalizer_params)
|
||||||
|
if activation_fn is not None:
|
||||||
|
outputs = activation_fn(outputs)
|
||||||
|
if normalizer_fn is not None and normalize_after_activation:
|
||||||
|
normalizer_params = normalizer_params or {}
|
||||||
|
outputs = normalizer_fn(outputs, **normalizer_params)
|
||||||
|
return outputs
|
||||||
|
|
||||||
|
class Vbn_double(object):
|
||||||
|
def __init__(self, x, epsilon=1e-5, scope=None):
|
||||||
|
shape = x.get_shape().as_list()
|
||||||
|
needs_reshape = len(shape) != 4
|
||||||
|
if needs_reshape:
|
||||||
|
orig_shape = shape
|
||||||
|
if len(shape) == 2:
|
||||||
|
if data_format == 'NCHW':
|
||||||
|
x = tf.reshape(x, [shape[0], shape[1], 0, 0])
|
||||||
|
else:
|
||||||
|
x = tf.reshape(x, [shape[0], 1, 1, shape[1]])
|
||||||
|
elif len(shape) == 1:
|
||||||
|
x = tf.reshape(x, [shape[0], 1, 1, 1])
|
||||||
|
else:
|
||||||
|
assert False, shape
|
||||||
|
shape = x.get_shape().as_list()
|
||||||
|
with tf.variable_scope(scope):
|
||||||
|
self.epsilon = epsilon
|
||||||
|
self.scope = scope
|
||||||
|
self.mean, self.var = tf.nn.moments(x, [0,2,3], \
|
||||||
|
keep_dims=True)
|
||||||
|
self.inv_std = tf.rsqrt(self.var + epsilon)
|
||||||
|
self.batch_size = int(x.get_shape()[0])
|
||||||
|
out = self._normalize(x, self.mean, self.inv_std)
|
||||||
|
if needs_reshape:
|
||||||
|
out = tf.reshape(out, orig_shape)
|
||||||
|
self.reference_output = out
|
||||||
|
|
||||||
|
def __call__(self, x):
|
||||||
|
shape = x.get_shape().as_list()
|
||||||
|
needs_reshape = len(shape) != 4
|
||||||
|
if needs_reshape:
|
||||||
|
orig_shape = shape
|
||||||
|
if len(shape) == 2:
|
||||||
|
if self.data_format == 'NCHW':
|
||||||
|
x = tf.reshape(x, [shape[0], shape[1], 0, 0])
|
||||||
|
else:
|
||||||
|
x = tf.reshape(x, [shape[0], 1, 1, shape[1]])
|
||||||
|
elif len(shape) == 1:
|
||||||
|
x = tf.reshape(x, [shape[0], 1, 1, 1])
|
||||||
|
else:
|
||||||
|
assert False, shape
|
||||||
|
with tf.variable_scope(self.scope, reuse=True):
|
||||||
|
out = self._normalize(x, self.mean, self.inv_std)
|
||||||
|
if needs_reshape:
|
||||||
|
out = tf.reshape(out, orig_shape)
|
||||||
|
return out
|
||||||
|
|
||||||
|
def _normalize(self, x, mean, inv_std):
|
||||||
|
shape = x.get_shape().as_list()
|
||||||
|
assert len(shape) == 4
|
||||||
|
gamma = tf.get_variable("gamma", [1,shape[1],1,1],
|
||||||
|
initializer=tf.constant_initializer(1.))
|
||||||
|
beta = tf.get_variable("beta", [1,shape[1],1,1],
|
||||||
|
initializer=tf.constant_initializer(0.))
|
||||||
|
coeff = gamma * inv_std
|
||||||
|
return (x * coeff) + (beta - mean * coeff)
|
||||||
|
|
||||||
|
@add_arg_scope
|
||||||
|
def vbn_double(ref_half, real_half, center=True, scale=True, epsilon=1e-5, \
|
||||||
|
data_format='NCHW', instance_norm=True, scope=None, \
|
||||||
|
reuse=None):
|
||||||
|
assert isinstance(epsilon, float)
|
||||||
|
shape = real_half.get_shape().as_list()
|
||||||
|
batch_size = int(real_half.get_shape()[0])
|
||||||
|
with tf.variable_scope(scope, 'VBN', reuse=reuse):
|
||||||
|
if data_format == 'NCHW':
|
||||||
|
if scale:
|
||||||
|
gamma = tf.get_variable("gamma", [1,shape[1],1,1],
|
||||||
|
initializer=tf.constant_initializer(1.))
|
||||||
|
if center:
|
||||||
|
beta = tf.get_variable("beta", [1,shape[1],1,1],
|
||||||
|
initializer=tf.constant_initializer(0.))
|
||||||
|
ref_mean, ref_var = tf.nn.moments(ref_half, [0,2,3], \
|
||||||
|
keep_dims=True)
|
||||||
|
else:
|
||||||
|
if scale:
|
||||||
|
gamma = tf.get_variable("gamma", [1,1,1,shape[-1]],
|
||||||
|
initializer=tf.constant_initializer(1.))
|
||||||
|
if center:
|
||||||
|
beta = tf.get_variable("beta", [1,1,1,shape[-1]],
|
||||||
|
initializer=tf.constant_initializer(0.))
|
||||||
|
ref_mean, ref_var = tf.nn.moments(ref_half, [0,1,2], \
|
||||||
|
keep_dims=True)
|
||||||
|
def _normalize(x, mean, var):
|
||||||
|
inv_std = tf.rsqrt(var + epsilon)
|
||||||
|
if scale:
|
||||||
|
coeff = inv_std * gamma
|
||||||
|
else:
|
||||||
|
coeff = inv_std
|
||||||
|
if center:
|
||||||
|
return (x * coeff) + (beta - mean * coeff)
|
||||||
|
else:
|
||||||
|
return (x - mean) * coeff
|
||||||
|
if instance_norm:
|
||||||
|
if data_format == 'NCHW':
|
||||||
|
real_mean, real_var = tf.nn.moments(real_half, [2,3], \
|
||||||
|
keep_dims=True)
|
||||||
|
else:
|
||||||
|
real_mean, real_var = tf.nn.moments(real_half, [1,2], \
|
||||||
|
keep_dims=True)
|
||||||
|
real_coeff = 1. / (batch_size + 1.)
|
||||||
|
ref_coeff = 1. - real_coeff
|
||||||
|
new_mean = real_coeff * real_mean + ref_coeff * ref_mean
|
||||||
|
new_var = real_coeff * real_var + ref_coeff * ref_var
|
||||||
|
ref_output = _normalize(ref_half, ref_mean, ref_var)
|
||||||
|
real_output = _normalize(real_half, new_mean, new_var)
|
||||||
|
else:
|
||||||
|
ref_output = _normalize(ref_half, ref_mean, ref_var)
|
||||||
|
real_output = _normalize(real_half, ref_mean, ref_var)
|
||||||
|
return ref_output, real_output
|
||||||
|
|
||||||
|
|
||||||
|
@add_arg_scope
|
||||||
|
def vbn_single(x, center=True, scale=True, \
|
||||||
|
epsilon=1e-5, data_format='NCHW', \
|
||||||
|
instance_norm=True, scope=None, \
|
||||||
|
reuse=None):
|
||||||
|
assert isinstance(epsilon, float)
|
||||||
|
shape = x.get_shape().as_list()
|
||||||
|
if shape[0] is None:
|
||||||
|
half_size = x.shape[0] // 2
|
||||||
|
else:
|
||||||
|
half_size = shape[0] // 2
|
||||||
|
needs_reshape = len(shape) != 4
|
||||||
|
if needs_reshape:
|
||||||
|
orig_shape = shape
|
||||||
|
if len(shape) == 2:
|
||||||
|
if data_format == 'NCHW':
|
||||||
|
x = tf.reshape(x, [shape[0], shape[1], 0, 0])
|
||||||
|
else:
|
||||||
|
x = tf.reshape(x, [shape[0], 1, 1, shape[1]])
|
||||||
|
elif len(shape) == 1:
|
||||||
|
x = tf.reshape(x, [shape[0], 1, 1, 1])
|
||||||
|
else:
|
||||||
|
assert False, shape
|
||||||
|
shape = x.get_shape().as_list()
|
||||||
|
batch_size = int(x.get_shape()[0])
|
||||||
|
with tf.variable_scope(scope, 'VBN', reuse=reuse):
|
||||||
|
ref_half = tf.slice(x, [0,0,0,0], [half_size, shape[1], \
|
||||||
|
shape[2], shape[3]])
|
||||||
|
if data_format == 'NCHW':
|
||||||
|
if scale:
|
||||||
|
gamma = tf.get_variable("gamma", [1,shape[1],1,1],
|
||||||
|
initializer=tf.constant_initializer(1.))
|
||||||
|
if center:
|
||||||
|
beta = tf.get_variable("beta", [1,shape[1],1,1],
|
||||||
|
initializer=tf.constant_initializer(0.))
|
||||||
|
ref_mean, ref_var = tf.nn.moments(ref_half, [0,2,3], \
|
||||||
|
keep_dims=True)
|
||||||
|
else:
|
||||||
|
if scale:
|
||||||
|
gamma = tf.get_variable("gamma", [1,1,1,shape[-1]],
|
||||||
|
initializer=tf.constant_initializer(1.))
|
||||||
|
if center:
|
||||||
|
beta = tf.get_variable("beta", [1,1,1,shape[-1]],
|
||||||
|
initializer=tf.constant_initializer(0.))
|
||||||
|
ref_mean, ref_var = tf.nn.moments(ref_half, [0,1,2], \
|
||||||
|
keep_dims=True)
|
||||||
|
def _normalize(x, mean, var):
|
||||||
|
inv_std = tf.rsqrt(var + epsilon)
|
||||||
|
if scale:
|
||||||
|
coeff = inv_std * gamma
|
||||||
|
else:
|
||||||
|
coeff = inv_std
|
||||||
|
if center:
|
||||||
|
return (x * coeff) + (beta - mean * coeff)
|
||||||
|
else:
|
||||||
|
return (x - mean) * coeff
|
||||||
|
if instance_norm:
|
||||||
|
real_half = tf.slice(x, [half_size,0,0,0], \
|
||||||
|
[half_size, shape[1], shape[2], shape[3]])
|
||||||
|
if data_format == 'NCHW':
|
||||||
|
real_mean, real_var = tf.nn.moments(real_half, [2,3], \
|
||||||
|
keep_dims=True)
|
||||||
|
else:
|
||||||
|
real_mean, real_var = tf.nn.moments(real_half, [1,2], \
|
||||||
|
keep_dims=True)
|
||||||
|
real_coeff = 1. / (batch_size + 1.)
|
||||||
|
ref_coeff = 1. - real_coeff
|
||||||
|
new_mean = real_coeff * real_mean + ref_coeff * ref_mean
|
||||||
|
new_var = real_coeff * real_var + ref_coeff * ref_var
|
||||||
|
ref_output = _normalize(ref_half, ref_mean, ref_var)
|
||||||
|
real_output = _normalize(real_half, new_mean, new_var)
|
||||||
|
return tf.concat([ref_output, real_output], axis=0)
|
||||||
|
else:
|
||||||
|
return _normalize(x, ref_mean, ref_var)
|
||||||
|
|
||||||
|
|
@ -0,0 +1,77 @@
|
||||||
|
import argparse
|
||||||
|
import numpy as np
|
||||||
|
import tensorflow as tf
|
||||||
|
from functools import partial
|
||||||
|
|
||||||
|
from utils import *
|
||||||
|
from generator import *
|
||||||
|
from queues import *
|
||||||
|
from YeNet import YeNet
|
||||||
|
|
||||||
|
parser = argparse.ArgumentParser(description='PyTorch implementation of YeNet')
|
||||||
|
parser.add_argument('train_cover_dir', type=str, metavar='PATH',
|
||||||
|
help='path of directory containing all ' +
|
||||||
|
'training cover images')
|
||||||
|
parser.add_argument('train_stego_dir', type=str, metavar='PATH',
|
||||||
|
help='path of directory containing all ' +
|
||||||
|
'training stego images or beta maps')
|
||||||
|
parser.add_argument('valid_cover_dir', type=str, metavar='PATH',
|
||||||
|
help='path of directory containing all ' +
|
||||||
|
'validation cover images')
|
||||||
|
parser.add_argument('valid_stego_dir', type=str, metavar='PATH',
|
||||||
|
help='path of directory containing all ' +
|
||||||
|
'validation stego images or beta maps')
|
||||||
|
parser.add_argument('--batch-size', type=int, default=32, metavar='N',
|
||||||
|
help='input batch size for training (default: 32)')
|
||||||
|
parser.add_argument('--test-batch-size', type=int, default=32, metavar='N',
|
||||||
|
help='input batch size for testing (default: 32)')
|
||||||
|
parser.add_argument('--epochs', type=int, default=1000, metavar='N',
|
||||||
|
help='number of epochs to train (default: 1000)')
|
||||||
|
parser.add_argument('--lr', type=float, default=4e-1, metavar='LR',
|
||||||
|
help='learning rate (default: 4e-1)')
|
||||||
|
parser.add_argument('--use-batch-norm', action='store_true', default=False,
|
||||||
|
help='use batch normalization after each activation,' +
|
||||||
|
' also disable pair constraint (default: False)')
|
||||||
|
parser.add_argument('--embed-otf', action='store_true', default=False,
|
||||||
|
help='use beta maps and embed on the fly instead' +
|
||||||
|
' of use stego images (default: False)')
|
||||||
|
parser.add_argument('--no-cuda', action='store_true', default=False,
|
||||||
|
help='disables CUDA training')
|
||||||
|
parser.add_argument('--gpu', type=int, default=0,
|
||||||
|
help='index of gpu used (default: 0)')
|
||||||
|
parser.add_argument('--seed', type=int, default=1, metavar='S',
|
||||||
|
help='random seed (default: 1)')
|
||||||
|
parser.add_argument('--log-interval', type=int, default=200, metavar='N',
|
||||||
|
help='how many batches to wait ' +
|
||||||
|
'before logging training status')
|
||||||
|
parser.add_argument('--log-path', type=str, default='logs/',
|
||||||
|
metavar='PATH', help='path to generated log file')
|
||||||
|
args = parser.parse_args()
|
||||||
|
|
||||||
|
import os
|
||||||
|
os.environ['CUDA_DEVICE_ORDER'] = 'PCI_BUS_ID'
|
||||||
|
os.environ['CUDA_VISIBLE_DEVICES'] = '' if args.no_cuda else str(args.gpu)
|
||||||
|
|
||||||
|
tf.set_random_seed(args.seed)
|
||||||
|
train_ds_size = len(glob(args.train_cover_dir + '/*')) * 2
|
||||||
|
if args.embed_otf:
|
||||||
|
train_gen = partial(gen_embedding_otf, args.train_cover_dir, \
|
||||||
|
args.train_stego_dir, args.use_batch_norm)
|
||||||
|
else:
|
||||||
|
train_gen = partial(gen_flip_and_rot, args.train_cover_dir, \
|
||||||
|
args.train_stego_dir, args.use_batch_norm)
|
||||||
|
|
||||||
|
valid_ds_size = len(glob(args.valid_cover_dir + '/*')) * 2
|
||||||
|
valid_gen = partial(gen_valid, args.valid_cover_dir, \
|
||||||
|
args.valid_stego_dir)
|
||||||
|
|
||||||
|
if valid_ds_size % 32 != 0:
|
||||||
|
raise ValueError("change batch size for validation")
|
||||||
|
|
||||||
|
optimizer = tf.train.AdadeltaOptimizer(args.lr)
|
||||||
|
|
||||||
|
|
||||||
|
train(YeNet, train_gen, valid_gen, args.batch_size, \
|
||||||
|
args.test_batch_size, valid_ds_size, \
|
||||||
|
optimizer, args.log_interval, train_ds_size, \
|
||||||
|
args.epochs * train_ds_size, train_ds_size, args.log_path, 8)
|
||||||
|
|
@ -0,0 +1,120 @@
|
||||||
|
import numpy as np
|
||||||
|
import tensorflow as tf
|
||||||
|
import threading
|
||||||
|
import h5py
|
||||||
|
import functools
|
||||||
|
|
||||||
|
def hdf5baseGen(filepath, thread_idx, n_threads):
|
||||||
|
with h5py.File(filepath, 'r') as f:
|
||||||
|
keys = f.keys()
|
||||||
|
nb_data = f[keys[0]].shape[0]
|
||||||
|
idx = thread_idx
|
||||||
|
while True:
|
||||||
|
yield [np.expand_dims(f[key][idx], 0) for key in keys]
|
||||||
|
idx = (idx + n_threads) % nb_data
|
||||||
|
|
||||||
|
class GeneratorRunner():
|
||||||
|
"""
|
||||||
|
This class manage a multithreaded queue filled with a generator
|
||||||
|
"""
|
||||||
|
def __init__(self, generator, capacity):
|
||||||
|
"""
|
||||||
|
inputs: generator feeding the data, must have thread_idx
|
||||||
|
as parameter (but the parameter may be not used)
|
||||||
|
"""
|
||||||
|
self.generator = generator
|
||||||
|
_input = generator(0,1).next()
|
||||||
|
if type(_input) is not list:
|
||||||
|
raise ValueError("generator doesn't return" \
|
||||||
|
"a list: %r" % type(_input))
|
||||||
|
input_batch_size = _input[0].shape[0]
|
||||||
|
if not all(_input[i].shape[0] == input_batch_size for i in range(len(_input))):
|
||||||
|
raise ValueError("all the inputs doesn't have " + \
|
||||||
|
"the same batch size," \
|
||||||
|
"the batch sizes are: %s" % [_input[i].shape[0] \
|
||||||
|
for i in range(len(_input))])
|
||||||
|
self.data = []
|
||||||
|
self.dtypes = []
|
||||||
|
self.shapes = []
|
||||||
|
for i in range(len(_input)):
|
||||||
|
self.shapes.append(_input[i].shape[1:])
|
||||||
|
self.dtypes.append(_input[i].dtype)
|
||||||
|
self.data.append(tf.placeholder(dtype=self.dtypes[i], \
|
||||||
|
shape=(input_batch_size,) + self.shapes[i]))
|
||||||
|
self.queue = tf.FIFOQueue(capacity, shapes=self.shapes, \
|
||||||
|
dtypes=self.dtypes)
|
||||||
|
self.enqueue_op = self.queue.enqueue_many(self.data)
|
||||||
|
self.close_queue_op = self.queue.close(cancel_pending_enqueues=True)
|
||||||
|
|
||||||
|
def get_batched_inputs(self, batch_size):
|
||||||
|
"""
|
||||||
|
Return tensors containing a batch of generated data
|
||||||
|
"""
|
||||||
|
batch = self.queue.dequeue_many(batch_size)
|
||||||
|
return batch
|
||||||
|
|
||||||
|
def thread_main(self, sess, thread_idx=0, n_threads=1):
|
||||||
|
try:
|
||||||
|
for data in self.generator(thread_idx, n_threads):
|
||||||
|
sess.run(self.enqueue_op, feed_dict={i: d \
|
||||||
|
for i, d in zip(self.data, data)})
|
||||||
|
if self.stop_threads:
|
||||||
|
return
|
||||||
|
except RuntimeError:
|
||||||
|
pass
|
||||||
|
except tf.errors.CancelledError:
|
||||||
|
pass
|
||||||
|
|
||||||
|
def start_threads(self, sess, n_threads=1):
|
||||||
|
self.stop_threads = False
|
||||||
|
self.threads = []
|
||||||
|
for n in range(n_threads):
|
||||||
|
t = threading.Thread(target=self.thread_main, args=(sess, n, n_threads))
|
||||||
|
t.daemon = True
|
||||||
|
t.start()
|
||||||
|
self.threads.append(t)
|
||||||
|
return self.threads
|
||||||
|
|
||||||
|
def stop_runner(self, sess):
|
||||||
|
self.stop_threads = True
|
||||||
|
# j = 0
|
||||||
|
# while np.any([t.is_alive() for t in self.threads]):
|
||||||
|
# j += 1
|
||||||
|
# if j % 100 = 0:
|
||||||
|
# print [t.is_alive() for t in self.threads]
|
||||||
|
sess.run(self.close_queue_op)
|
||||||
|
|
||||||
|
def queueSelection(runners, sel, batch_size):
|
||||||
|
selection_queue = tf.FIFOQueue.from_list(sel, [r.queue for r in runners])
|
||||||
|
return selection_queue.dequeue_many(batch_size)
|
||||||
|
|
||||||
|
def doubleQueue(runner1, runner2, is_runner1, batch_size1, batch_size2):
|
||||||
|
return tf.cond(is_runner1, lambda: runner1.queue.dequeue_many(batch_size1), \
|
||||||
|
lambda: runner2.queue.dequeue_many(batch_size2))
|
||||||
|
|
||||||
|
if __name__ == '__main__':
|
||||||
|
def randomGen(img_size, enqueue_batch_size, thread_idx, n_threads):
|
||||||
|
while True:
|
||||||
|
batch_of_1_channel_imgs = np.random.rand(enqueue_batch_size, \
|
||||||
|
img_size, img_size, 1)
|
||||||
|
batch_of_labels = np.random.randint(0,11,enqueue_batch_size)
|
||||||
|
return [batch_of_1_channel_imgs, batch_of_labels]
|
||||||
|
|
||||||
|
TRAIN_BATCH_SIZE = 64
|
||||||
|
VALID_BATCH_SIZE = 10
|
||||||
|
train_runner = GeneratorRunner(functool.partial(randomGen, \
|
||||||
|
(128, 10)), TRAIN_BATCH_SIZE * 10)
|
||||||
|
valid_runner = GeneratorRunner(functool.partial(randomGen, \
|
||||||
|
(128, 10)), VALID_BATCH_SIZE * 10)
|
||||||
|
is_training = tf.Variable(True)
|
||||||
|
batch_size = tf.Variable(TRAIN_BATCH_SIZE)
|
||||||
|
enable_training_op = tf.group(tf.assign(is_training, True), \
|
||||||
|
tf.assign(batch_size, TRAIN_BATCH_SIZE))
|
||||||
|
disable_training_op = tf.group(tf.assign(is_training, False), \
|
||||||
|
tf.assign(batch_size, VALID_BATCH_SIZE))
|
||||||
|
img_batch, label_batch = queueSelection([valid_runner, train_runner], \
|
||||||
|
tf.cast(is_training, tf.int32), \
|
||||||
|
batch_size)
|
||||||
|
# img_batch, label_batch = doubleQueue(train_runner, valid_runner, \
|
||||||
|
# is_training, TRAIN_BATCH_SIZE, \
|
||||||
|
# VALID_BATCH_SIZE)
|
||||||
|
|
@ -0,0 +1,9 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Thu Aug 8 14:33:16 2019
|
||||||
|
|
||||||
|
@author: Lee
|
||||||
|
"""
|
||||||
|
import numpy as np
|
||||||
|
SRM_Kernels = np.load('SRM_Kernels.npy')
|
||||||
|
print(SRM_Kernels[:1])
|
||||||
|
|
@ -0,0 +1,9 @@
|
||||||
|
# -*- coding: utf-8 -*-
|
||||||
|
"""
|
||||||
|
Created on Wed Sep 4 15:24:05 2019
|
||||||
|
|
||||||
|
@author: Lee
|
||||||
|
"""
|
||||||
|
|
||||||
|
import torch
|
||||||
|
print(torch.rand(5))
|
||||||
|
|
@ -0,0 +1,298 @@
|
||||||
|
import tensorflow as tf
|
||||||
|
import numpy as np
|
||||||
|
import sys
|
||||||
|
import time
|
||||||
|
from glob import glob
|
||||||
|
from functools import partial
|
||||||
|
import os
|
||||||
|
from os.path import expanduser
|
||||||
|
home = expanduser("~")
|
||||||
|
user = home.split('/')[-1]
|
||||||
|
sys.path.append(home + '/tflib/')
|
||||||
|
from queues import *
|
||||||
|
from generator import *
|
||||||
|
|
||||||
|
def optimistic_restore(session, save_file, \
|
||||||
|
graph=tf.get_default_graph()):
|
||||||
|
reader = tf.train.NewCheckpointReader(save_file)
|
||||||
|
saved_shapes = reader.get_variable_to_shape_map()
|
||||||
|
var_names = sorted([(var.name, var.name.split(':')[0]) for var in tf.global_variables()
|
||||||
|
if var.name.split(':')[0] in saved_shapes])
|
||||||
|
restore_vars = []
|
||||||
|
for var_name, saved_var_name in var_names:
|
||||||
|
curr_var = graph.get_tensor_by_name(var_name)
|
||||||
|
var_shape = curr_var.get_shape().as_list()
|
||||||
|
if var_shape == saved_shapes[saved_var_name]:
|
||||||
|
restore_vars.append(curr_var)
|
||||||
|
opt_saver = tf.train.Saver(restore_vars)
|
||||||
|
opt_saver.restore(session, save_file)
|
||||||
|
|
||||||
|
class average_summary(object):
|
||||||
|
def __init__(self, variable, name, num_iterations):
|
||||||
|
self.sum_variable = tf.get_variable(name, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0.), \
|
||||||
|
dtype='float32', \
|
||||||
|
trainable=False, \
|
||||||
|
collections=[tf.GraphKeys.LOCAL_VARIABLES])
|
||||||
|
with tf.control_dependencies([variable]):
|
||||||
|
self.increment_op = tf.assign_add(self.sum_variable, variable)
|
||||||
|
self.mean_variable = self.sum_variable / float(num_iterations)
|
||||||
|
self.summary = tf.summary.scalar(name, self.mean_variable)
|
||||||
|
with tf.control_dependencies([self.summary]):
|
||||||
|
self.reset_variable_op = tf.assign(self.sum_variable, 0)
|
||||||
|
|
||||||
|
def add_summary(self, sess, writer, step):
|
||||||
|
s, _ = sess.run([self.summary, self.reset_variable_op])
|
||||||
|
writer.add_summary(s, step)
|
||||||
|
|
||||||
|
class Model(object):
|
||||||
|
def __init__(self, is_training=None, data_format='NCHW'):
|
||||||
|
self.data_format = data_format
|
||||||
|
if is_training is None:
|
||||||
|
self.is_training = tf.get_variable('is_training', dtype=tf.bool, \
|
||||||
|
initializer=tf.constant_initializer(True), \
|
||||||
|
trainable=False)
|
||||||
|
else:
|
||||||
|
self.is_training = is_training
|
||||||
|
|
||||||
|
def _build_model(self, inputs):
|
||||||
|
raise NotImplementedError('Here is your model definition')
|
||||||
|
|
||||||
|
def _build_losses(self, labels):
|
||||||
|
self.labels = tf.cast(labels, tf.int64)
|
||||||
|
with tf.variable_scope('loss'):
|
||||||
|
oh = tf.one_hot(self.labels, 2)
|
||||||
|
self.loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits( \
|
||||||
|
labels=oh, logits=self.outputs))
|
||||||
|
with tf.variable_scope('accuracy'):
|
||||||
|
am = tf.argmax(self.outputs, 1)
|
||||||
|
equal = tf.equal(am, self.labels)
|
||||||
|
self.accuracy = tf.reduce_mean(tf.cast(equal, tf.float32))
|
||||||
|
return self.loss, self.accuracy
|
||||||
|
|
||||||
|
def train(model_class, train_gen, valid_gen, train_batch_size, \
|
||||||
|
valid_batch_size, valid_ds_size, optimizer, \
|
||||||
|
train_interval, valid_interval, max_iter, \
|
||||||
|
save_interval, log_path, num_runner_threads=1, \
|
||||||
|
load_path=None):
|
||||||
|
tf.reset_default_graph()
|
||||||
|
train_runner = GeneratorRunner(train_gen, train_batch_size * 10)
|
||||||
|
valid_runner = GeneratorRunner(valid_gen, valid_batch_size * 10)
|
||||||
|
is_training = tf.get_variable('is_training', dtype=tf.bool, \
|
||||||
|
initializer=True, trainable=False)
|
||||||
|
if train_batch_size == valid_batch_size:
|
||||||
|
batch_size = train_batch_size
|
||||||
|
disable_training_op = tf.assign(is_training, False)
|
||||||
|
enable_training_op = tf.assign(is_training, True)
|
||||||
|
else:
|
||||||
|
batch_size = tf.get_variable('batch_size', dtype=tf.int32, \
|
||||||
|
initializer=train_batch_size, \
|
||||||
|
trainable=False, \
|
||||||
|
collections=[tf.GraphKeys.LOCAL_VARIABLES])
|
||||||
|
disable_training_op = tf.group(tf.assign(is_training, False), \
|
||||||
|
tf.assign(batch_size, valid_batch_size))
|
||||||
|
enable_training_op = tf.group(tf.assign(is_training, True), \
|
||||||
|
tf.assign(batch_size, train_batch_size))
|
||||||
|
img_batch, label_batch = queueSelection([valid_runner, train_runner], \
|
||||||
|
tf.cast(is_training, tf.int32), \
|
||||||
|
batch_size)
|
||||||
|
model = model_class(is_training, 'NCHW')
|
||||||
|
model._build_model(img_batch)
|
||||||
|
loss, accuracy = model._build_losses(label_batch)
|
||||||
|
regularization_losses = tf.get_collection(
|
||||||
|
tf.GraphKeys.REGULARIZATION_LOSSES)
|
||||||
|
regularized_loss = tf.add_n([loss] + regularization_losses)
|
||||||
|
train_loss_s = average_summary(loss, 'train_loss', train_interval)
|
||||||
|
train_accuracy_s = average_summary(accuracy, 'train_accuracy', \
|
||||||
|
train_interval)
|
||||||
|
valid_loss_s = average_summary(loss, 'valid_loss', \
|
||||||
|
float(valid_ds_size) / float(valid_batch_size))
|
||||||
|
valid_accuracy_s = average_summary(accuracy, 'valid_accuracy', \
|
||||||
|
float(valid_ds_size) / float(valid_batch_size))
|
||||||
|
global_step = tf.get_variable('global_step', dtype=tf.int32, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0), \
|
||||||
|
trainable=False)
|
||||||
|
minimize_op = optimizer.minimize(regularized_loss, global_step)
|
||||||
|
train_op = tf.group(minimize_op, train_loss_s.increment_op, \
|
||||||
|
train_accuracy_s.increment_op)
|
||||||
|
increment_valid = tf.group(valid_loss_s.increment_op, \
|
||||||
|
valid_accuracy_s.increment_op)
|
||||||
|
init_op = tf.group(tf.global_variables_initializer(), \
|
||||||
|
tf.local_variables_initializer())
|
||||||
|
saver = tf.train.Saver(max_to_keep=10000)
|
||||||
|
with tf.Session() as sess:
|
||||||
|
sess.run(init_op)
|
||||||
|
if load_path is not None:
|
||||||
|
loader = tf.train.Saver(reshape=True)
|
||||||
|
loader.restore(sess, load_path)
|
||||||
|
train_runner.start_threads(sess, num_runner_threads)
|
||||||
|
valid_runner.start_threads(sess, 1)
|
||||||
|
writer = tf.summary.FileWriter(log_path + '/LogFile/', \
|
||||||
|
sess.graph)
|
||||||
|
start = sess.run(global_step)
|
||||||
|
sess.run(disable_training_op)
|
||||||
|
sess.run([valid_loss_s.reset_variable_op, \
|
||||||
|
valid_accuracy_s.reset_variable_op, \
|
||||||
|
train_loss_s.reset_variable_op, \
|
||||||
|
train_accuracy_s.reset_variable_op])
|
||||||
|
_time = time.time()
|
||||||
|
for j in range(0, valid_ds_size, valid_batch_size):
|
||||||
|
sess.run([increment_valid])
|
||||||
|
_acc_val = sess.run(valid_accuracy_s.mean_variable)
|
||||||
|
print "validation:", _acc_val, " | ", \
|
||||||
|
"duration:", time.time() - _time, \
|
||||||
|
"seconds long"
|
||||||
|
valid_accuracy_s.add_summary(sess, writer, start)
|
||||||
|
valid_loss_s.add_summary(sess, writer, start)
|
||||||
|
sess.run(enable_training_op)
|
||||||
|
print valid_interval
|
||||||
|
for i in xrange(start+1, max_iter+1):
|
||||||
|
sess.run(train_op)
|
||||||
|
if i % train_interval == 0:
|
||||||
|
train_loss_s.add_summary(sess, writer, i)
|
||||||
|
train_accuracy_s.add_summary(sess, writer, i)
|
||||||
|
if i % valid_interval == 0:
|
||||||
|
sess.run(disable_training_op)
|
||||||
|
for j in range(0, valid_ds_size, valid_batch_size):
|
||||||
|
sess.run([increment_valid])
|
||||||
|
valid_loss_s.add_summary(sess, writer, i)
|
||||||
|
valid_accuracy_s.add_summary(sess, writer, i)
|
||||||
|
sess.run(enable_training_op)
|
||||||
|
if i % save_interval == 0:
|
||||||
|
saver.save(sess, log_path + '/Model_' + str(i) + '.ckpt')
|
||||||
|
|
||||||
|
def test_dataset(model_class, gen, batch_size, ds_size, load_path):
|
||||||
|
tf.reset_default_graph()
|
||||||
|
runner = GeneratorRunner(gen, batch_size * 10)
|
||||||
|
img_batch, label_batch = runner.get_batched_inputs(batch_size)
|
||||||
|
model = model_class(False, 'NCHW')
|
||||||
|
model._build_model(img_batch)
|
||||||
|
loss, accuracy = model._build_losses(label_batch)
|
||||||
|
loss_summary = average_summary(loss, 'loss', \
|
||||||
|
float(ds_size) / float(batch_size))
|
||||||
|
accuracy_summary = average_summary(accuracy, 'accuracy', \
|
||||||
|
float(ds_size) / float(batch_size))
|
||||||
|
increment_op = tf.group(loss_summary.increment_op, \
|
||||||
|
accuracy_summary.increment_op)
|
||||||
|
global_step = tf.get_variable('global_step', dtype=tf.int32, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0), \
|
||||||
|
trainable=False)
|
||||||
|
init_op = tf.group(tf.global_variables_initializer(), \
|
||||||
|
tf.local_variables_initializer())
|
||||||
|
saver = tf.train.Saver(max_to_keep=10000)
|
||||||
|
with tf.Session() as sess:
|
||||||
|
sess.run(init_op)
|
||||||
|
saver.restore(sess, load_path)
|
||||||
|
runner.start_threads(sess, 1)
|
||||||
|
for j in range(0, ds_size, batch_size):
|
||||||
|
sess.run(increment_op)
|
||||||
|
mean_loss, mean_accuracy = sess.run([loss_summary.mean_variable ,\
|
||||||
|
accuracy_summary.mean_variable])
|
||||||
|
print "Accuracy:", mean_accuracy, " | Loss:", mean_loss
|
||||||
|
|
||||||
|
def find_best(model_class, valid_gen, test_gen, valid_batch_size, \
|
||||||
|
test_batch_size, valid_ds_size, test_ds_size, load_paths):
|
||||||
|
tf.reset_default_graph()
|
||||||
|
valid_runner = GeneratorRunner(valid_gen, valid_batch_size * 30)
|
||||||
|
img_batch, label_batch = valid_runner.get_batched_inputs(valid_batch_size)
|
||||||
|
model = model_class(False, 'NCHW')
|
||||||
|
model._build_model(img_batch)
|
||||||
|
loss, accuracy = model._build_losses(label_batch)
|
||||||
|
loss_summary = average_summary(loss, 'loss', \
|
||||||
|
float(valid_ds_size) \
|
||||||
|
/ float(valid_batch_size))
|
||||||
|
accuracy_summary = average_summary(accuracy, 'accuracy', \
|
||||||
|
float(valid_ds_size) \
|
||||||
|
/ float(valid_batch_size))
|
||||||
|
increment_op = tf.group(loss_summary.increment_op, \
|
||||||
|
accuracy_summary.increment_op)
|
||||||
|
global_step = tf.get_variable('global_step', dtype=tf.int32, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0), \
|
||||||
|
trainable=False)
|
||||||
|
init_op = tf.group(tf.global_variables_initializer(), \
|
||||||
|
tf.local_variables_initializer())
|
||||||
|
saver = tf.train.Saver(max_to_keep=10000)
|
||||||
|
accuracy_arr = []
|
||||||
|
loss_arr = []
|
||||||
|
print "validation"
|
||||||
|
for load_path in load_paths:
|
||||||
|
with tf.Session() as sess:
|
||||||
|
sess.run(init_op)
|
||||||
|
saver.restore(sess, load_path)
|
||||||
|
valid_runner.start_threads(sess, 1)
|
||||||
|
_time = time.time()
|
||||||
|
for j in range(0, valid_ds_size, valid_batch_size):
|
||||||
|
sess.run(increment_op)
|
||||||
|
mean_loss, mean_accuracy = sess.run([loss_summary.mean_variable ,\
|
||||||
|
accuracy_summary.mean_variable])
|
||||||
|
accuracy_arr.append(mean_accuracy)
|
||||||
|
loss_arr.append(mean_loss)
|
||||||
|
print load_path
|
||||||
|
print "Accuracy:", accuracy_arr[-1], "| Loss:", loss_arr[-1], \
|
||||||
|
"in", time.time() - _time, "seconds."
|
||||||
|
argmax = np.argmax(accuracy_arr)
|
||||||
|
print "best savestate:", load_paths[argmax], "with", \
|
||||||
|
accuracy_arr[argmax], "accuracy and", loss_arr[argmax], \
|
||||||
|
"loss on validation"
|
||||||
|
print "test:"
|
||||||
|
test_dataset(model_class, test_gen, test_batch_size, test_ds_size, \
|
||||||
|
load_paths[argmax])
|
||||||
|
return argmax, accuracy_arr, loss_arr
|
||||||
|
|
||||||
|
|
||||||
|
def extract_stats_outputs(model_class, gen, batch_size, ds_size, load_path):
|
||||||
|
tf.reset_default_graph()
|
||||||
|
runner = GeneratorRunner(gen, batch_size * 10)
|
||||||
|
img_batch, label_batch = runner.get_batched_inputs(batch_size)
|
||||||
|
model = model_class(False, 'NCHW')
|
||||||
|
model._build_model(img_batch)
|
||||||
|
global_step = tf.get_variable('global_step', dtype=tf.int32, shape=[], \
|
||||||
|
initializer=tf.constant_initializer(0), \
|
||||||
|
trainable=False)
|
||||||
|
init_op = tf.group(tf.global_variables_initializer(), \
|
||||||
|
tf.local_variables_initializer())
|
||||||
|
saver = tf.train.Saver(max_to_keep=10000)
|
||||||
|
stats_outputs_arr = np.empty([ds_size, \
|
||||||
|
model.stats_outputs.get_shape().as_list()[1]])
|
||||||
|
with tf.Session() as sess:
|
||||||
|
sess.run(init_op)
|
||||||
|
saver.restore(sess, load_path)
|
||||||
|
runner.start_threads(sess, 1)
|
||||||
|
for j in range(0, ds_size, batch_size):
|
||||||
|
stats_outputs_arr[j:j+batch_size] = sess.run(model.stats_outputs)
|
||||||
|
return stats_outputs_arr
|
||||||
|
|
||||||
|
def stats_outputs_all_datasets(model_class, ds_head_dir, payload, \
|
||||||
|
algorithm, load_path, save_dir):
|
||||||
|
if not os.path.exists(save_dir):
|
||||||
|
os.makedirs(save_dir + '/')
|
||||||
|
payload_str = ''.join(str(payload).strip('.'))
|
||||||
|
train_ds_size = len(glob(ds_head_dir + '/train/cover/*'))
|
||||||
|
valid_ds_size = len(glob(ds_head_dir + '/valid/cover/*'))
|
||||||
|
test_ds_size = len(glob(ds_head_dir + '/test/cover/*'))
|
||||||
|
train_gen = partial(gen_all_flip_and_rot, ds_head_dir + \
|
||||||
|
'/train/cover/', ds_head_dir + '/train/' + \
|
||||||
|
algorithm + '/payload' + payload_str + '/stego/')
|
||||||
|
valid_gen = partial(gen_valid, ds_head_dir + '/valid/cover/', \
|
||||||
|
ds_head_dir + '/valid/' + algorithm + \
|
||||||
|
'/payload' + payload_str + '/stego/')
|
||||||
|
test_gen = partial(gen_valid, ds_head_dir + '/test/cover/', \
|
||||||
|
ds_head_dir + '/test/' + algorithm + \
|
||||||
|
'/payload' + payload_str + '/stego/')
|
||||||
|
print "train..."
|
||||||
|
stats_outputs = extract_stats_outputs(model_class, train_gen, 16, \
|
||||||
|
train_ds_size * 2 * 4 * 2, \
|
||||||
|
load_path)
|
||||||
|
stats_shape = stats_outputs.shape
|
||||||
|
stats_outputs = stats_outputs.reshape(train_ds_size, 2, 4, \
|
||||||
|
2, stats_shape[-1])
|
||||||
|
stats_outputs = np.transpose(stats_outputs, axes=[0,3,2,1,4])
|
||||||
|
np.save(save_dir + '/train.npy', stats_outputs)
|
||||||
|
print "validation..."
|
||||||
|
stats_outputs = extract_stats_outputs(model_class, valid_gen, 16, \
|
||||||
|
valid_ds_size * 2, load_path)
|
||||||
|
np.save(save_dir + '/valid.npy', stats_outputs)
|
||||||
|
print "test..."
|
||||||
|
stats_outputs = extract_stats_outputs(model_class, test_gen, 16, \
|
||||||
|
test_ds_size * 2, load_path)
|
||||||
|
np.save(save_dir + '/test.npy', stats_outputs)
|
||||||
Binary file not shown.
Binary file not shown.
Binary file not shown.
Some files were not shown because too many files have changed in this diff Show More
Loading…
Reference in New Issue